Identification of polymorphisms in melanocortin 1 receptor gene and their association with coat color variants in Black Bengal goat
Abstract
In domestic goats, coat color is genetically determined as an important character and has an economic value. The aim of this study was to identify polymorphisms in the entire coding regions of the melanocortin 1 receptor (MC1R) gene and their possible associations with coat color variants of the Black Bengal goat (BBG) of Bangladesh. Forty blood samples representing five distinct color variants of BBG (Solid Black, Toggenburg, Dutch Belt, White and Brown) were collected from Bangladesh Livestock Research Institute, Savar, Dhaka, Bangladesh. Two PCR amplicons that harbored entire coding region were sequenced both forward and reverse directions. Multiple sequence alignment identified four single nucleotide polymorphisms (SNPs) based on Capra hircus reference sequence (MT186778.1). Among them, the c.183C>T (p.61A) polymorphism was identified as synonymous type and the remaining three were non-synonymous in nature (c.676A>G, c.748G>T, and c.801C>G), causing amino acid substitutions as p.K226E, p.V250F, and p.C267W, respectively. Twelve haplotypes were defined by four substitutions where four haplotypes (Hap6, Hap7, Hap8, and Hap9) were found white-color goat-specific involving c.801C>G mutation that might be associated with white coat color phenotype of BBG. The UPGMA phylogenetic tree revealed two separate clusters, one of which included all color variants of BBG and major domestic goat populations (Capra hircus) from various countries. The haplotype sharing features suggest that the coat color phenotypes of BBG were not solely attributable to the MC1R gene. However, more detailed investigation with large samples from diverse populations across the country is required to draw a precise conclusion.
INTRODUCTION
Coat color is a clearly identifiable characteristic feature for any breed or variety of domestic mammalian and avian species. Coat color distinguishes animals’ phenotypes and is a qualitative trait controlled by only a single or few genes [1]. Many breeders include coat color as an important element of their animal selection criteria. Coat color evolved as an adaptive trait for a particular coat hue [2,3] and farmers also choose to keep a specific goat color for better market price. Goats with black coats are preferred in Bangladesh over those with white or brown due to the high demand for their meat and skin [4]. Uncovering the genetic basis of coat color leads to the formation of specific herds or flocks for coat color preference markets [5]. Around the world, a broad variety of coat color phenotypes in goats have been described. The two main loci that regulate the relative amount of eumelanin and pheomelanin production in cells are Extension (E) and Agouti (A) [6]. These loci show evidence of epistatic interactions in several animals. Dominant alleles at the E locus induce black pigmentation whereas recessive alleles boost the formation of pheomelanin, which is responsible for red, yellow, and light pigmentation. The melanocortin 1 receptor (MC1R) protein, which is encoded by the E locus, is a member of the G protein coupled receptor family and possesses seven transmembrane domains. It binds to melanocyte-stimulating hormone (MSH) and stimulates the synthesis of eumelanin [7]. Conversely, the agouti signaling protein (ASIP), which acts as an MC1R antagonist and affects pigmentation, is encoded by the A locus. It prevents contact with MSH-receptor and triggers for shifting pigment-type from eumelanin to pheomelanin [8, 9]. Previous study showed that mutations in the MC1R gene affect the coat colors of several mammals, including mice [7], cattle [10,11,12] pigs [13], horses [14], sheep [15] and goat [16], with gain of function mutations producing black/dark coat color and loss of function mutations producing red/yellow or white coat color.
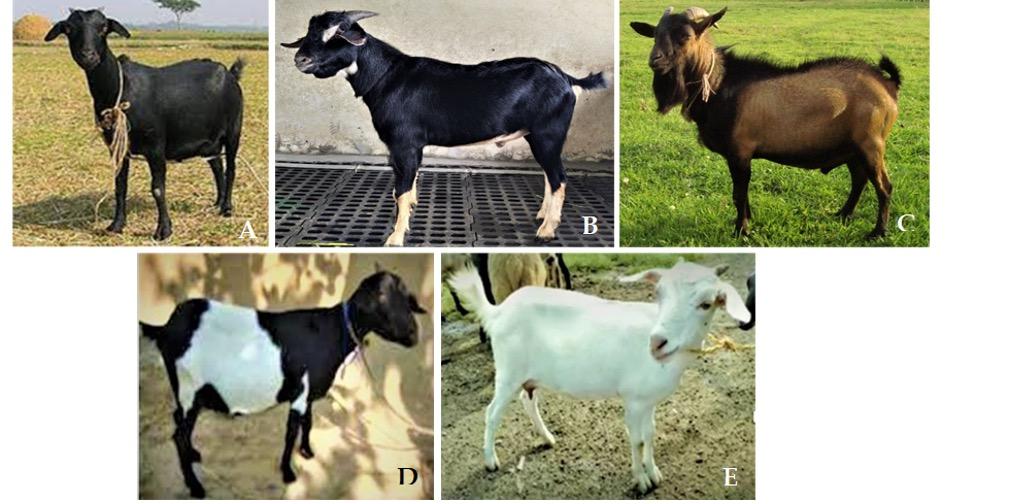
Goat is an important livestock species under integrated farming system of Bangladesh. Bangladesh has 26.774 million goats [17] that belongs to one breed popularly known as the Black Bengal goat (BBG) and accounts for approximately 90% of the overall goat population [18]. BBG is an economically important animal genetic resource with potential contribution to the sustainable agricultural system. The BBG is found all around Bangladesh, West Bengal, Bihar, Assam, and Odisha [19]. This dwarf-type goat breed has a worldwide reputation for their early sexual maturity, higher fertility rate, high prolificacy, adaptability to hot and humid conditions, superior meat and skin quality [20,21, 22]. BBG exhibits diverse types of coat color phenotypes. Chowdhury 2002 [4] reported predominantly Solid black (amounting 69% of the total goat population), Toggenburg (13%), Brown (5%), Solid White (4%) and Dutch belt (9%) colored goat. However, the underlying genetic cause for coat color variation in BBG has not yet been disclosed. In this study, these five color variants of BBG were included in order to sequence entire coding region of MC1R gene and to detect genetic polymorphisms in the MC1R gene and their possible association with coat color variants in BBG of Bangladesh.
MATERIALS AND METHODS
Ethical statement
The study was conducted in accordance with the protocol approved by the Animal Experimentation Ethics Committee of Bangladesh Livestock Research Institute, Savar, Dhaka (Approval no.: AEEC/BLRI00104/2023).
Blood sampling and DNA extraction
A total of 40 blood samples representing five distinct color variants of BBG were collected from Bangladesh Livestock Research Institute (BLRI) Headquarters Savar, Dhaka except Brown Bengal that was collected from BLRI regional station, Naikhongchhari, Bandarban district (Figure 1). Blood samples were taken from the jugular vein using venoject tubes coated with EDTA as anticoagulant. Precautions were made to take samples from mature and unrelated animals. DNA extraction was performed by using AddPrep Genomic DNA Extraction Kit (ADD BIO INC., South Korea) from the whole blood according to the manufacturer’s instructions with some modifications. After DNA extraction, the concentration and purity of isolated DNA was measured by Nanodrop spectrophotometer (Model DN2000, Thermo Fisher Scientific, USA) and was stored at -20°C for further use.
PCR amplification and gel electrophoresis
Two pairs of primers having 567 and 641 bp amplicon size previously reported by Guan et al. 2021 [23] were used in this study for PCR amplification. Primer sequence information, fragment length and optimized annealing temperature is shown in Table 1. Annealing temperature was optimized using gradient PCR protocol. PCR amplification of target fragments of MC1R gene was performed using TECHNE thermocycler (Bibby Scientific, UK) in a 20 μl reaction volume comprising 10 μl (2x conc.) Add Taq Master mix (20mM Tris-HCl [pH-8.8], 100mM KCl, 0.2% Triton® X-100, 4mM MgCl2, protein stabilizer, sediment, loading dye and 0.5mM each of dATP, dCTP, dGTP and dTTP), 2 μl of each primer (10 pmol/μl), 2 μl of genomic DNA (~50 ng/μl) and 4 μl double distilled water. The thermal profile consists of initial denaturation at 95°C for 10 min, 30 cycles of denaturation at 95°C for 30 sec, annealing at 62°C for 30 sec and extension at 72°C for 1 min, and final extension at 72°C for 10 min. Five (5) μl PCR product was used for agarose gel electrophoresis (2.0%) in order to confirm the availability of amplified fragments of target gene. Agarose gel was prepared using hazard free safe DNA gel stain. The image of the gel with electrophoresed DNA was then documented using digital gel documentation system (GDS-200, Sunil-Bio INC., South Korea) and marked for identification of each individual sample.
Table 1. Primers are used in PCR amplification.
Sequencing of the MC1R gene fragment
The PCR products were purified using purification kit (Macrogen, South Korea) and sequencing of the amplicon was performed both in forward and reverse directions by 3500 ×l Genetic Analyzer (Applied Biosystems, Foster City. CA, USA) from a commercial sequencing service provider (Wuhan Tianyi Huayu Gene Technology, Wuhan, China).
Statistical analysis
The generated sequences of MC1R gene were edited and aligned using bioinformatic tools BioEdit v.7.2.5 [24], ClustalW [25] and MEGA v.10.2 [26]. The resultant DNA fragments were aligned to the reference sequence of Capra hircus (MT186778.1) to obtain the entire coding sequence (CDS). Detection of mutation types were performed using NCBI supported ORF finder tool and MEGA software. The identified MC1R variants and their respective positions corresponding to the protein sequence were assigned by GeneDoc v.2.6 software. The genetic diversity parameters, such as haplotype diversity (Hd), nucleotide diversity (Pi), number of haplotypes (H) and number of segregating sites (S) were estimated using the DnaSP v.6.12 [27]. Haplotype file generation for further haplotype network analysis was also performed using the aforesaid software. A median-Joining (MJ) network was constructed using NETWORK v.5.0.0.1 [28] to uncover the genetic relationships among MC1R haplotypes in color variants of BBG. Using MEGA v.10.2, a UPGMA phylogenetic tree was built based on maximum composite likelihood and 10,000 bootstrap replications. For phylogenetic analysis, coding sequences of different domestic goats (Capra hircus) populations namely Koraput (OK160060.1), Ganjam (OK160058.1), Tai-hang Native-black goat (KT247426.1), Murciano-Granadina (MT186775.1), Camosciata delle Alpi (FM212940.1). In addition, MC1R coding sequences of Pig (KR865958.1), Sheep (NM_001282528.1), Horse (NM_001114534.1), Buffalo (MF421522.1) and Zebu cattle (MG373763.1) were included as out groups.
RESULTS
Identification of single nucleotide polymorphism
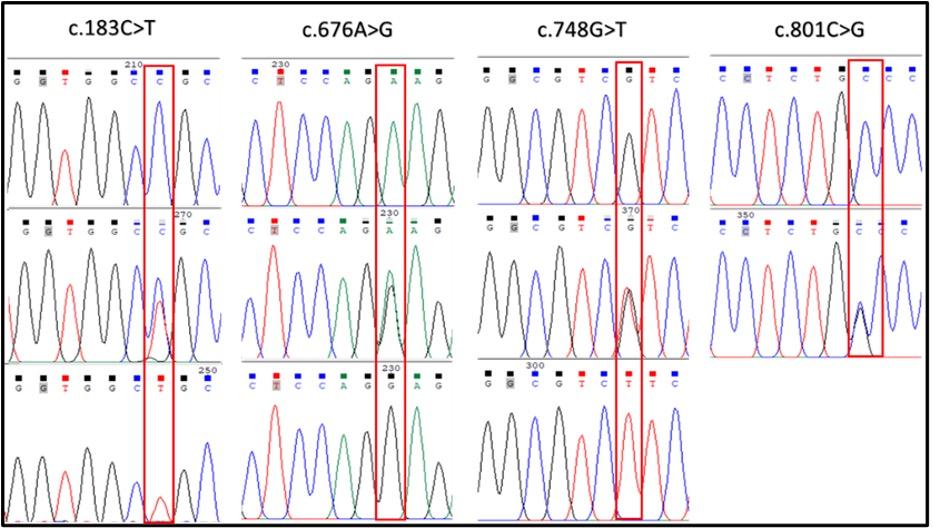
A total of 37 DNA samples comprising 9 Black Bengal, 9 Toggenburg, 8 Dutch Belt, 8 White and 3 brown colored goats were successfully amplified the MC1R gene fragments. When the resulting BBG CDS sequences were aligned, a total of four SNPs were identified (Table 2, Figure 2). Among them, one identified as synonymous type, c.183C>T (p.A61A), while the remaining three were nonsynonymous in nature, c.676A>G, c.748G>T, and c.801C>G, resulting amino acid changes as p.K226E, p.V250F, and p.C267W, respectively (Table 2, Figure 3). However, no polymorphism was detected in the BBG color variant with a brown hue. The goat MC1R gene's CDS region encodes for a 317 amino acid protein (Figure 3).
Table 2. Identified polymorphisms in the MC1R gene of BBG of Bangladesh

Genetic diversity parameters estimation
Table 3 showed the intra-population diversity measures in four color variants of BBG. The highest number of segregating sites (4) and haplotypes (7) was observed in White and Toggenburg goats while the lowest number of haplotypes (5) was observed in Solid Black goat. The highest haplotype diversity was observed in white goats (0.879±0.08) and the lowest was in Dutch belt goats (0.778±0.14). Consequently, the highest nucleotide diversity (Pi) was observed for white colored goat (0.0017±0.0002) and was lowest in Dutch belt goat (0.0012±0.0003).
Table 3. Diversity parameters of MC1R gene in different coat color variants of BBG
Haplotypes and phylogenetic analysis
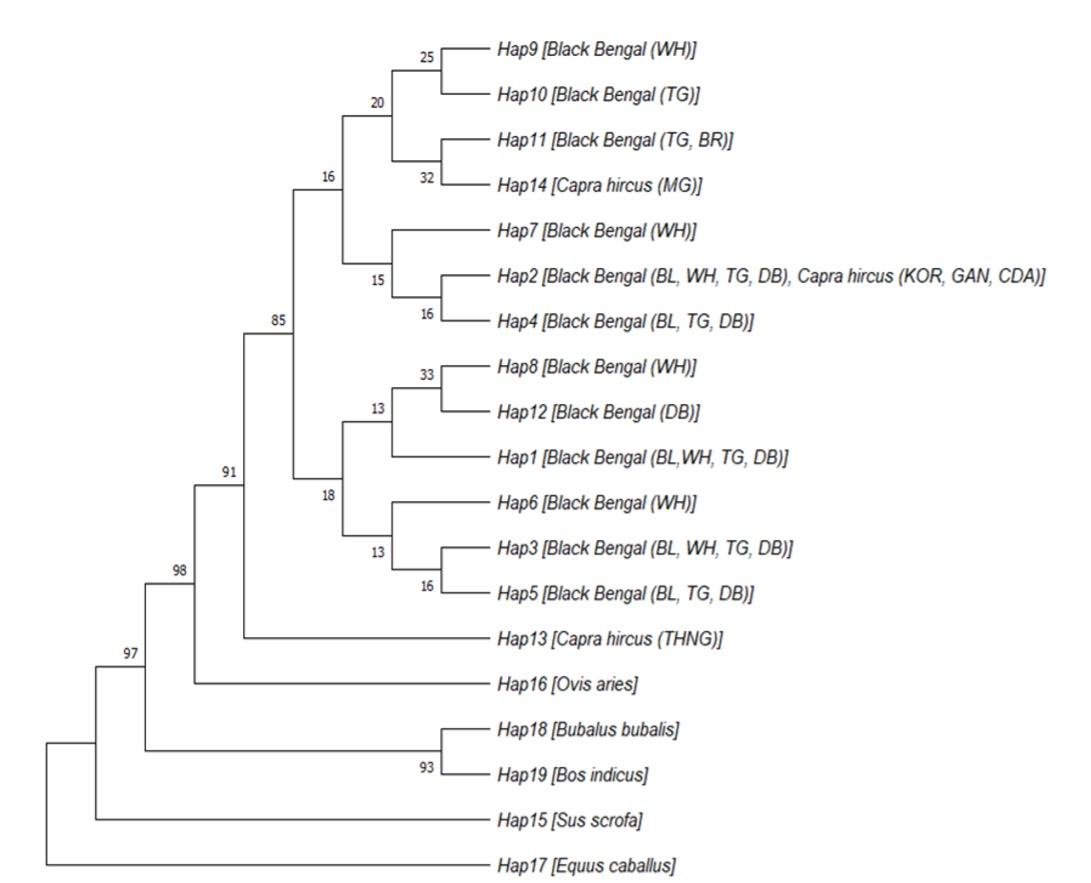
Table 4 enlisted the haplotypes and their frequencies based on four polymorphisms identified in different color variants of BBG. The majority of the recorded phenotypes were represented by the haplotype H1 (CGTC), which was followed by H2 (CATC, 27.0%) and H3 (CGGC, 21.6%). The remaining haplotypes were represented by comparatively fewer animals (ranging from 4 to 12) and is shown in Table 4. Interestingly, the haplotypes H6, H7, H8, and H9 were specific to white goats, whereas H10 and H12 belonged to Toggenburg and Dutch belt goats, respectively. According to the median-joining network analysis (Figure not shown), the haplotypes H1 (CGTC), H2 (CATC), H3 (CGGC), and H12 (TGTC) were closely related to each other, while H4 (TATC), H5 (TGGC), H6 (CGGG), H7 (CATG), H8 (TGTG), and H9 (CAGC) were separated by two mutation steps (c.676A>G and c.183C>T; c.183C>T and c.748G>T; c.748G>T and c.801C>G; c.676A>G and c.801C>G; c.183C>T and c.801C>G; c.748G>T and c.676A>G, respectively). In addition, H10 (CAGG) and H11 (TAGC) were separated by three mutational steps (c.676A>G, c.748G>T, c.801C>G and c.676A>G, c.183C>T, c.748G>T, respectively) from central H1 (CGTC) haplotype. A UPGMA phylogenetic tree was constructed based on maximum composite likelihood method (Figure 4). As expected, all out grouped animals showed distinctly separate clusters while the Capra hircus goat population formed two separate clusters (Figure 4). One cluster included haplotypes (Hap2, Hap4, Hap7, Hap9, Hap10, Hap11 and Hap14) of all color variants of BBG and most of the Capra hircus reference samples which depicted their close relatedness among them. The other haplotypes (Hap1, Hap3, Hap5, Hap6, Hap8 and Hap12) made a separate cluster including only Black Bengal color variants that specify them as a distinct group particularly for MC1R gene variants. Notably, Hap13 was separated from all goat haplotypes with a node that signifies different genetic architecture of THNG goat even as a member of Capra hircus.
Table 4. Constructed haplotypes and their frequencies in different coat color variants of BBG
DISCUSSION
Substantial research has been carried out to elucidate the genetic basis of coat color inheritance in different farm animals utilizing candidate genes’ information including MC1R. Here, the current research provides the molecular underpinnings of coat color inheritance in BBG of Bangladesh. According to reports, black and white colorations in animals are linked to mutations in the MC1R gene, and the black versus white coat color variation in goats might be actively influenced by the Extension locus [29]. The expression level of MC1R was significantly higher (P<0.01) in the skin of black Cashmere goats than that of white counterparts. However, the missense mutation on MC1R is not the only causal agent that can result in the white phenotype in Cashmere goats and there are additional potential causative agents that might affect coat coloration in Cashmere goats [30].
To best of our knowledge, this is the first report on MC1R gene’s polymorphism detection in BBG of Bangladesh. The identified four mutations in BBG (c.183C>T, c.676A>G, c.748G>T, and c.801C>G) are in accordance with the findings of earlier studies in various goat breeds around the world. Maharani et al. 2019 [31] detected three of the aforesaid mutations (c.676A>G, c.748G>T, and c.801C>G) in the coding region of MC1R gene of 10 Indonesian goat populations namely Gembrong, Senduro, Ettawa Grade, Boerawa, Boerka, Kosta, Samosir, Muara, Boer, and Kacang breeds. Guan et al. 2021 [23] reported the c.748G>T and c.801C>G mutations, while Fontanesi et al. 2009 [16] and Beretti et al. 2007 [32] found only the c.801C>G mutation in the Murciano-Granadina (MG) goat breed having solid black or solid brown phenotypes and support the present findings. A missense mutation c.676A>G was identified by Ganbold et al. 2019 [29] in Mongolian goats, Javanmard et al. 2015 [5] in Saanen White goats, Maharani et al. 2019 [31] in Ettawa grade goats, and Wu et al. 2006 [33] in Shannan White goats. Further, the silent mutation c.183C>T (p.61A) was reported by Ganbold et al. 2009 [29] in Mongolian goats and Fontanesi et al. 2009 [16] in Girgentana, Derivata di Siria, and Maltese goat breeds. All of these previously identified polymorphisms were exactly similar to this finding.
In this study, indispensable effects of MC1R gene mutations were not noticed on coat color variations of BBG of Bangladesh. The identified c.183C>T, c.676A>G, c.748G>T mutations were present in all of the four colored varieties of BBG and shared by major haplotypes. Whereas c.801C>G mutation was found to be unique for white Bengal goat and containing a higher frequency of mutated allele G (80%) than that of other color variants which indicated a significant association of this mutation with white coloration of BBG. However, this result contradicted the findings of Fontanesi et al. 2009 [16] and Guan et al. 2021 [23] who confirmed that the coat color of MG goats having solid black or solid brown is fully explained by MC1R c.801C>G genotype. According to Ganbold et al. 2009 [29], the c.676A>G (p. K226E) missense mutation showed a significantly greater AG and GG genotype frequencies in white (χ2=39.1; d.f. = 4; p<0.0001), while the AA genotypes were present in both red and black colored goat. For c.676A>G mutation, Wu et al. 2006 [33] found a higher frequency of GG genotype for majority of the investigated animals having complete white phenotypes, while only Boer (red-headed white) goats carry AA genotype. They also highlighted the G allele mutation may result in a loss of function mutation associated with complete white goat coat colors. However, Maharani et al. 2019 [31] confirmed the SNP g.676A>G of MC1R gene with Hardy-Weinberg equilibrium (P>0.05) indicating no strong selection in favor of any genotype regarding the coat color in the investigated goat breeds. Genotype and allele frequency are breed and population specific and therefore, allelic frequency differed largely from one population to another even for the same mutation.
The haplotype diversity (Hd) and nucleotide diversity (Pi) estimation allows researchers to assess the genetic variability within population [34]. In this study diversity measures suggest that white and Toggenburg goats possessed higher polymorphism than predominant Black and Dutch belt. Mongolian goat breeds having three different coat colors as black, white and red where the highest nucleotide diversity was observed in white animals [29] and is consistent to the current findings. Genetic diversity based on MC1R coding region sequences showed high haplotype diversity (0.85±0.05) and nucleotide diversity (0.002) in Creole sheep with White, Black, Brown, Dark gray and Light gray phenotypes [35] that support this study. Xi et al. 2012 [36] identified a total of nine SNPs, including four SNPs in the 5′-UTR and five SNPs in the CDS of MC1R gene and revealed high genetic diversity in gayal (Bos frontalis). According to Guo et al. 2010 [37], nucleotide diversity (Pi) in the strains of Hebei chickens (0.0047–0.0052) was significantly higher than that of Hy-Line Brown (0.0024) or Lohmann Brown (0.0043), pinpointing the greater polymorphisms in the MC1R gene, especially in Hebei chicken, which is associated with its rich plumage color diversity. However, all of the above-stated findings across the species support the present study. Taken together, genetic diversity measures are breed- or population-specific and differ largely from population to population, even within the same breed, due to selection practices and breeding methods.
Haplotypes are unique sequences of bases over a region of the genome. A total of twelve MC1R gene haplotypes were defined by four polymorphic sites in BBG. Haplotype H1, H2, H3, H4 and H5 were distributed among almost all the BBG goat color varieties. Whereas haplotype H6, H7, H8 and H9 were unique for only white goats might have a significant association with white phenotype. Seven MC1R haplotypes and an even higher number of ASIP haplotypes were observed in different goat populations [16, 29, 38]. According to Ganbold et al. 2019 [29] unique H5 (TGAGA) may have some relationship with black uniform which is incomparable to this study due to structural difference in the constructed haplotype. Eight haplotypes were constructed in caprine MC1R gene, and an association study revealed that haplotypes CCGG, TCGG, and CCTC were linked to white colored animals [39]. Fontanesi et al. 2009 [16] identified six haplotypes in the coding sequence of MC1R gene from six different goat breeds having different coat color and patterns where p.C267W allele derived haplotype was found in every black Murciano-Granadina goat and was absent in the brown ones. The haplotype AATGT in MC1R is uniquely associated with black coat color in Minxian Black-fur breed [40]. According to UPGMA phylogenetic tree, the Capra hircus goat population formed two distinct clusters in Black Bengal goat samples representing the existence of two separate lineages for MC1R gene.
CONCLUSION
A total of four SNPs were identified in the entire coding sequences of MC1R gene of BBG, among them the c.801C>G mutation was found to be unique for White Bengal goats. The highest haplotype and nucleotide diversity in the white colored goat pinpointing the presence of higher polymorphisms than that of predominant Solid black, Toggenburg and Dutch belt goats. A total of twelve MC1R gene haplotypes were defined by four polymorphic sites where major haplotypes H1, H2, H3 and H4 shared by all color variants of BBG. Taken together, this study suggests that the MC1R locus is probably not the only sole contributor for coat color features in BBG. The genetic components or interactions between loci that control the pigmentation of coat colors also need to be elucidated in BBG for making a precise conclusion.
ACKNOWLEDGEMENT
The author is grateful to the Goat Production Research Division, BLRI for providing valuable blood samples as well as financial support from the Black Bengal Goat Conservation and Development Research Project (Project ID: 224289000), BLRI for carrying out this research.
AUTHOR CONTRIBUTIONS
SA and MSAB conceptualized and designed the experiment. MAM and SA performed wet lab experimentation and analyzed data. MFAH and NHD collected phenotypic data and blood samples from goat farms. SA contributed to drafting the article. SA is responsible for financial support. MMH and MSAB contributed to revising the draft and made it up to the final stage.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Brooks S. Molecular Genetics of coat color: it is more than just skin deep. In: Khatib H (ed). Molecular and Quantitative Genetics. Wiley Blackwell, Madison, WI, USA, 2015, pp 187-195.
- [2]Banerjee AK, Animut G, et al. Selection and breeding strategies for increased productivity of goats in Ethiopia. In: Proceedings of a conference held at Debub University, Awassa, Ethiopia, 2000: 70-79
- [3]Manzi M, Rutagwenda T, et al. Phenotypic characterization of goats raised under traditional husbandry systems in Bugesera and Nyagatare districts of Rwanda. J Anim Vet Adv. 2011; 10(24): 3297-3302.
- [4]Chowdhury SA. Goat: Our natural resource and development opportunities. In: Proceedings of the Workshop on Poverty Alleviation Through Goat Production: National Program. Bangladesh Livestock Research Institute, Savar, Dhaka, Bangladesh. 2002: 1-51.
- [5]Javanmard A, Arafnajad B, et al. Polymorphisms in melanocortin receptor 1 gene in goat breeds: a window for coat color controlling mechanism. Iran J Appl Anim Sci. 2015; 5(4): 889-895.
- [6]Searle AG. Pigmentation and Inheritance: Comparative genetics of coat colour in mammals. Academic Press, New York, 1968, pp 667-678.
- [7]Robbins LS, Nadeau JH, et al. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 1993; 72(6): 827-834.
- [8]Lu D, Willard D, et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994; 371: 799-802.
- [9]Ollmann MM, Lamoreux ML, et al. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998; 12(3): 316-330.
- [10]Klungland H, Vage DI, et al. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 1995; 6: 636-639.
- [11]Joerg H, Fries HR, et al. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm Genome. 1996; 7(4): 317-318.
- [12]Rouzaud F, Martin J, et al. A first genotyping assay of French cattle breeds based on a new allele of the extension gene encoding the melanocortin-1 receptor (Mc1r). Genet Sel Evol. 2000; 32(5): 511-520.
- [13]Kijas JMH, Wales R, et al. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998; 150: 1177-1185.
- [14]Marklund L, Moller MJ, et al. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MCIR) is associated with the chestnut coat color in horses. Mamm Genome. 1996; 7: 895-899.
- [15]Våge DI, Klungland H, et al. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm Genome. 1999; 10(1): 39-43.
- [16]Fontanesi L, Beretti F, et al. Missense and nonsense mutations in melanocortin 1 receptor (MC1R) gene of different goat breeds: association with red and black coat color phenotypes but with unexpected evidences. BMC Genet. 2009; 10(1): 1-12.
- [17]BBS. Bangladesh Bureau of Statistics, Government of the People’s Republic of Bangladesh, 2022, pp 198.
- [18]Siddiky NA. Sustainable Goat Farming for Livelihood Improvement in South Asia. SAARC Agriculture Center, Dhaka-1215, Bangladesh, 2017, pp 190.
- [19]Mason IL. A world dictionary of breeds, types, and varieties of livestock, supplement no. 8, Commonwealth Agricultural Bureaux, 1969.
- [20]Devendra C, Burns M. Goat Production in the Tropics. Tech. Commun. Common. Bur. Anim. Breed. Genet., Commonwealth Agricultural Bureaux, England. 1983; Viii: 183.
- [21]Husain SS, Horst P, et al. Study on the growth performance of Black Bengal goats in different periods. Small Rumin Res. 1996; 21(3): 165-171.
- [22]Amin MR, Husain SS, et al. Reproductive peculiarities and litter weight in different genetic groups of Black Bengal does. Asian-Australas J Anim Sci. 2001; 14(3): 297-301.
- [23]Guan D, Martínez A, et al. Detecting the footprint of selection on the genomes of Murciano‐Granadina goats. Anim Genet. 2021; 52(5): 683-693.
- [24]Hall TA. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Sym. Ser. 1999; 41: 95-98.
- [25]Larkin MA, Blackshields G, et al. Clustal W and Clustal X version 2.0. J Bioinfor. 2007; 23(21): 2947-2948.
- [26]Kumar S, Stecher G, et al. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018; 35(6): 1547.
- [27]Rozas J, Ferrer-Mata A, et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol Biol Evol. 2017; 34: 3299-3302.
- [28]Bandelt HJ, Forster P, et al. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999; 16: 37-48.
- [29]Ganbold O, Manjula P, et al. Sequence characterization and polymorphism of melanocortin 1 receptor gene in some goat breeds with different coat color of Mongolia. Asian-Australas J Anim Sci. 2019; 32(7): 939.
- [30]Li J, Chen W, et al. Differential expression of MC1R gene in Liaoning Cashmere goats with different coat colors. Anim Biotechnol. 2019; 30(3): 273-278.
- [31]Maharani D, Elieser S, et al. Allelic and genotypic distribution in single nucleotide polymorphism (SNP) G. 676A>G of melanocortin-1 receptor (MC1R) gene in Indonesian goat breeds. Iran J Appl Anim Sci. 2019; 9(4): 687-692.
- [32]Beretti F, Finocchiaro R, et al. Analysis of the melanocortin receptor 1 (MC1R) gene in Sicilian goat breeds. Ital J Anim Sci. 2007; 6(sup1.): 46-46.
- [33]Wu ZL, Li XL, et al. The red head and neck of Boer goats may be controlled by the recessive allele of the MC1R gene. Anim Res. 2006; 55(4): 313-322.
- [34]Nei M. Molecular evolutionary genetics. Columbia University Press, New York, 1987, pp 512.
- [35]Hepp D, Gonçalves GL, et al. Identification of the e allele at the Extension locus (MC1R) in Brazilian Creole sheep and its role in wool color variation. Genet Mol Res. 2012; 11(3): 2997-3006.
- [36]Xi D, Liu Q, et al. Nucleotide diversity of the melanocortin 1 receptor gene (MC1R) in the gayal (Bos frontalis). Mol Biol Rep. 2012; 39: 7293-7301.
- [37]Guo XL, Li XL, et al. Genetic variation of chicken MC1R gene in different plumage color populations. Br Poult Sci. 2010; 51(6): 734-739.
- [38]Adefenwa MA, Peters SO, et al. Identification of single nucleotide polymorphisms in the agouti signaling protein (ASIP) gene in some goat breeds in tropical and temperate climates. Mol Biol Rep. 2013; 40: 4447-4457.
- [39]Venkatesh KM, Mishra C, et al. novel heterozygote allele in caprine melanocortin 1 receptor (MC1R) gene: an association with heat stress traits. Trop Anim Health Prod. 2023; 55(2): 68.
- [40]Yang GL, Fu DL, et al. Mutations in MC1R gene determine black coat color phenotype in Chinese sheep. Sci World J. 2013: 675382.