Determination of multi-drug resistance profile of isolated Gram-positive and Gram-negative bacteria from clinical pus samples in Bangladesh
Abstract
Antimicrobial resistance (AMR) has been a very concerning issue with consistent rise on a global scale. The rapid spread of drug-resistant pathogenic bacteria is a serious public health concern in both developed and developing countries, including Bangladesh. This study aimed to determine the prevalence of antibiotic resistant pathogenic bacteria and their multi-drug resistance (MDR) rate in pus samples. A total of 891 pus positive samples were collected from Tangail, Bangladesh. The resulting bacterial isolates were confirmed by biochemical tests and gram staining to classify bacterial species into two large groups. Antimicrobial susceptibility tests were performed for the identified bacterial isolates using the disk diffusion method. Escherichia coli exhibited the highest resistance level (98.92%, n=92). Also, Pseudomonas spp. displayed a substantial resistance pattern at 92.66% (n = 341), Proteus spp. at 91.58% (n = 87), Klebsiella spp. at 87.5% (n = 56), and Acinetobacter spp. exhibiting complete resistance of 100% (n =6). The cumulative MDR trend for Gram-negative bacteria was significant (92.98%, n = 583). Conversely, Gram-positive bacteria demonstrated a robust resistance pattern as well. Streptococcus spp. displayed resistance in 66.66% (n = 2) of cases, and Enterococcus faecalis exhibited resistance in 92.23% (n = 95) of instances. While Staphylococcus aureus showed a high resistance level with 95.06% (n = 77) of isolates. The overall drug-resistant pattern for Gram-positive bacteria was substantial (87.5%, n = 231). As per the findings of this study, bacteria frequently encountered demonstrate a concerning prevalence of MDR, posing a significant challenge to public health. The outcomes of this research may contribute valuable insights for formulating evidence-based treatment strategies and underscore the critical need for early identification of drug-resistant bacteria. Finally, this imperative step may hold the potential to mitigate the disease burden effectively.
INTRODUCTION
Infections, primarily caused by bacteria, play a pivotal role in the onset and advancement of diseases. When pathogens enter into the body through various entry points such as the skin, surgical wounds, mucous membranes, respiratory tract, and intraperitoneal injection, profound infections can ensue [1]. In the presence of bacterial or fungal infections, inflammation triggers the secretion of a fluid known as pus. The formation of pus is not limited to pus-forming microorganisms; it can also involve various non-pus-forming entities like fungi, viruses, protozoa, and others. Additionally, pyrogenic bacteria, when combined with deceased tissue, cells, and protein-enriched serum, can contribute to pus formation. It may manifest in diverse colors, including white, yellow, green, and brown [1-3]. The predominant bacterial strains found among pus samples are Pseudomonas spp., Proteus spp., E. coli, Klebsiella spp., Acinetobacter spp., and Enterobacter spp., which are Gram-negative. On the other hand, Gram-positive bacteria are Enterococcus faecalis, Staphylococcus aureus, Staphylococcus spp., Streptococcus spp., respectively [4]. Severe infections create a wet, warm, and nutritive environment favorable to microbial colonization, proliferation, and infection [5]. Pathogenic bacterial infections are important public health issues, and a considerable number of people die each day as a result of avoidable and treatable illnesses [6, 7].
Antibiotic resistance is developed among microorganisms through enzymatic degradation, modifications of bacterial surfaces target proteins, inactivation of a drug, folate metabolism, ribosome function, active efflux of a drug and changes to the permeability of cell membranes [8, 9]. Resistance emerges organically over time but misuse or overuse with extensive doses of unprescribed medication may contribute to antibiotic resistance in humans [10].
Multi-drug resistant bacteria (MDR) are more common globally, particularly in Bangladesh [11-14]. It is already declared as a public health hazard by the World Health Organization (WHO), and many governments have been advised to adopt an action plan to address the impending catastrophe [15, 16]. Based on current projections, drug-resistant illnesses were directly responsible for 1.27 million fatalities worldwide in 2019. There could be up to 10 million deaths a year by 2050. Whereas annually antibiotic-resistant organisms are indebted for at least 2 million diseases and 23,000 fatalities in the USA. Antimicrobial resistance (AMR) could drive 24 million more people into extreme poverty in the next ten years and remove 3.4 trillion USD from GDP yearly if left unchecked. By 2050, the world economy might lose more than 6 trillion USD annually as a result of AMR, which accounts for almost 4% of years global GDP [17].
Numerous studies have consistently shown that pus samples exhibit a higher concentration of bacterial isolates compared to various other clinical samples [3, 5, 18]. Thus, the objective of the current investigation is to assess the AMR profile of pathogenic microorganisms and to present an overview of the MDR pattern within pus samples obtained from infectious patients in two distinct localities in Bangladesh.
MATERIALS AND METHODS
Study areas and time frame
A total of 891 patients were enrolled in this study from January 2018 to March 2022. Patients with different body parts of infection including skin surface and inside, surgical incisions, burns, superficial and soft tissue infections, breast abscesses, and diabetic feet [19, 20] were collected by the Lab Zone, and Hormone Center of Tangail districts in Bangladesh. All the enrolled patients have provided their explicit consent for the participation in this study.
Ethical approval of the study
This study was approved by the Institutional Review Board Committee members from the Department of Biochemistry and Molecular Biology, Mawlana Bhashani Science and Technology University (No. MBSTU/BMB/TEST/6/2022/155).
Culture of Bacterial isolates from pus samples
Swab samples were inoculated into a variety of sterilized agar media, including Nutrient agar, Blood sheep agar, and MacConkey agar media (Oxoid, Basingstoke, Hampshire, UK) [21, 22]. Following proper incubation (at 370C for overnight growth), the culture plates were meticulously observed for the evidence of desired bacterial growth.
Identification of bacterial isolates
The bacterial colony size, shape, and color were the basic indicators for the initial identification of isolated bacteria. Gram staining techniques and various biochemical assays such as Klinger’s Iron Agar (KIA) test (Oxoid) include lactose fermenter Acid slant /Acid (A/A), Gas production and H2S production, Motility Indole Urea (MIU) test (Oxoid), Citrate, Catalase, Urease test (Oxoid) were performed to identify isolated organisms [23].
Antibiotic susceptibility testing
Antibiogram test using Mueller-Hinton agar media was conducted according to the Kirby-Bauers disk diffusion method. The susceptibility test was evaluated by measuring the zone of inhibition in the diameter of the millimeter with a transparent plastic scale following the recommendations of the Clinical and Laboratory Standards Institute (CLSI) guidelines. Antibiotics were divided into 13 different classes including fluoroquinolones (Levofloxacin 5µg, Ciprofloxacin 5µg), tetracyclines (Tetracycline 30µg), macrolides (Azithromycin 15µg), carbapenems (Meropenem 10µg), aminoglycosides (Amikacin 30µg, Gentamicin 10μg), lincosamide (Clindamycin 2µg), Oxazolidinone (Linezolid 30µg), nitrofuran (Nitrofurantoin 300μg) antibiotics, cephalosporins (Ceftriaxone 30µg, Ceftazidime 30μg , Cefixime 5µg, Cephalexin 2µg, Cephradine 30μg, Cefuroxime 30μg), Azole antifungals (Fluconazole 8µg), polymyxin (Colistin), and sulfonamides (Sulfamethoxazole/Trimethoprim 2.5µg), and penicillin (Penicillin-G 10 IU, Amoxicillin 30 µg).
Statistical analysis
The statistical analysis, and data visualization were performed using IBM Inc.'s SPSS 20.0 and Microsoft Excel 2016 (Chicago, USA). The trends for AMR and MDR were identified using descriptive statistics. For each age group, sex, and infection etiology, descriptive data such as frequency distribution and percentage were calculated.
RESULTS
Distribution of the bacterial population in pus samples
This investigation found 10 different bacterial species from where 70% (n = 627) samples and 30% (n = 264) were gram-positive bacteria. Gram-negative bacteria included Pseudomonas spp., Proteus spp., E. coli, Klebsiella spp., Acinetobacter spp., Enterobacter spp. and gram-positive bacterial isolates were Enterococcus faecalis, Staphylococcus aureus, Staphylococcus spp., and Streptococcus spp. (Table 1).
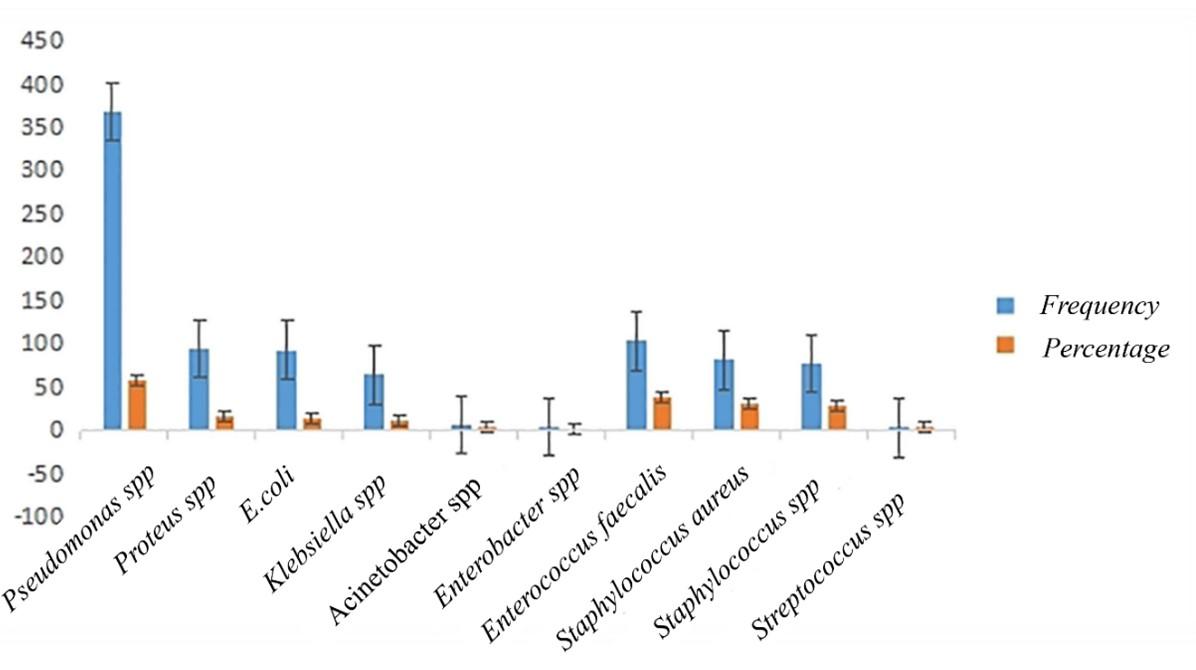
Among gram-negative isolates, Pseudomonas spp. was the most predominant 58.69% (n = 368), followed by Proteus spp. 15.15% (n = 95), E. coli 14.83% (n = 93), Klebsiella spp. 10.20% (n = 64) and Acinetobacter spp. (n=6, 0.95%) (Figure 1). Gram-positive isolates, with Enterococcus faecalis being the predominant species at 39.02% (n=103), were observed in this study. Further, Staphylococcus aureus accounted for 30.68% (n=81), followed by Staphylococcus spp. at 29.16% (n=77), and Streptococcus spp. at 1.13% (n=3) (Table 1).
Table 1. Bacterial isolates from different pus infections were stained using gram staining.
Identification of bacterial isolates according to different age and sex categories
Specific bacterial isolates into different age groups were categorized and shown in Table 2. The first group comprised of infants in an age range of (0-2 years), where we have found three bacterial isolates including Pseudomonas spp. (n=1), Klebsiealla spp. (n=1), and Staphylococcus aureus (n=1). The second group was children (3-16 years), where bacterial isolates Pseudomonas spp. (n=83, 41.09%) were present mostly, followed by Proteus spp. (n=66, 32.67%). In Young Adults (17-30 years), Pseudomonas spp. (n=79, 36.41%) and Enterococcus faecalis (n=61, 28.11%) were found at a high rate. Also, in middle-aged adults (31-45 years) and old adults (≥45 years), the highest number of bacterial isolates were Pseudomonas spp. (n=90, 48.13%), and (n=115, 40.78%), respectively (Table 2).
In this study, about male (n= 640, 71.8%) and female (n=251, 28.1%) from a total of 891 pus samples were identified by sex categories. From young adults age rage (17-30), identified (n=217) the highest number of bacterial isolates where male contain 7 types of bacteria where female contain 5 types as same male (Table 3).
Table 2. Frequency and percentage of predominant bacterial isolates according to age range.
Table 3. Frequency and percentage of bacterial isolates according to age range and sex.
Antibiotic susceptibility profile of Gram-negative bacteria in pus samples
We conducted a drug susceptibility test on Gram-negative bacterial isolates, evaluating their resistance to 20 antibiotics spanning thirteen different classes. These classes include aminoglycosides, cephalosporins, fluoroquinolones, tetracyclines, carbapenems, lincosamides, oxazolidinones, penicillins, sulfonamides, macrolides, nitrofurantoin, polymyxins, and azole antifungals.
Among Gram-negative bacteria, E. coli demonstrated the highest resistance, with 100% (n=93) against amoxicillin (AMC). Proteus spp. exhibited substantial resistance, with 92.6% (n=88) against ceftazidime (CAZ) and cefixime (CFM). Pseudomonas spp. displayed notable resistance, with 79.6% (n=293) against cefixime (CFM), while Klebsiella spp. showed resistance, with 87.5% (n=56) against cephalexin (CT) and cefepime (FEP). Acinetobacter spp. demonstrated pervasive resistance, with 100% (n=6) against sixteen antibiotics, excluding amikacin (AK), gentamicin (GEN), meropenem (MEM), and levofloxacin (LE). Enterobacter spp. exhibited resistance, with 100% (n=1), against 8 out of 20 antibiotics (Table 4).
Table 4. Antibiotic susceptibility pattern of Gram-negative bacterial isolates in pus samples.
Antibiotic susceptibility profile of Gram-positive bacteria in pus samples
Enterococcus faecalis exhibited complete resistance (n=103, 100%) to eight out of twenty antibiotics, including Amoxicillin, Ceftriaxone, Cefuroxime, Ciprofloxacin, Clindamycin, Cefepime, Fluconazole, and Sulphamethoxazole. In contrast, Streptococcus spp. showed utmost resistance (n=3, 100%) to Cephradine, Cefixime, and Cefuroxime. Cephalexin demonstrated the highest resistance (n=67, 82.7%) in Staphylococcus aureus. Notably, 71.4% of Staphylococcus spp. displayed resistance to clindamycin (Table 5).
Table 5. Antibiotic susceptibility pattern of Gram-positive bacterial isolates in pus samples.
MDR pattern of Gram-negative and Gram-positive bacteria in pus samples
From 627 gram-negative bacterial isolates, 92.98%(n=583) were identified as MDR. Among them Pseudomonas spp. (92.66%), Proteus spp. (91.58%), E. coli (98.92%), and Klebsiella spp. (87.5%) were found in most of the cases (Table 6). In the case of gram-positive bacteria, 87.5% (n=231) of the 264 isolates have shown their resistance in multiple antibiotics (Table 6). Where, Enterococcus faecalis (92. 23%) and Staphylococcus aureus (95.06%) demonstrated a high prevalence of drug resistance (Table 6).
Table 6. MDR pattern of Gram-negative and Gram-positive bacteria isolated from pus samples.
DISCUSSION
Despite advancements in surgical techniques and the use of antibiotic prophylaxis, pus samples still pose a significant health risk to the public. Due to AMR, postoperative infection prevention is still a major concern for medical professionals worldwide [24, 25]. One of the primary reasons for increasing bacterial resistance against antibiotics is the inappropriate use of antibiotics to treat bacterial illnesses. Our research emphasized the identification of antibiotic-resistant and multidrug-resistant bacteria commonly associated with various diseases in pus samples. It also examined the prevalence of antibiotic resistance and the effectiveness of specific antibiotics against both Gram-negative and Gram-positive bacteria, with a particular focus on Tangail districts. Age, Sex, and Gram staining were considered independent variables. Gram-negative bacteria isolated from the pus sample exhibited a slightly higher percentage of MDR pattern than Gram -positive bacteria.
According to our findings, the most common Gram-negative isolates were Pseudomonas spp., followed by Proteus spp., E. coli, Klebsiella spp., Acinetobacter spp., and Enterobacter spp. and in Gram-positive isolates were Enterococcus faecalis, Staphylococcus aureus, Staphylococcus spp., and Streptococcus spp. Similar findings were observed by a previous study [26].
The Gram-negative bacteria Pseudomonas spp. showed the highest sensitivity to Amikacin in 292(79.3%) cases and resistance in Cefixime to 293(79.6%) cases. For Proteus spp., Amikacin and Meropenem were sensitive in 90 (94.7%) cases, whereas Cefixime showed resistance in 88(n=92.6%) cases. Meropenem displayed 100% sensitivity in E. coli, while Linozolid and Clindamycin exhibited resistance in 92(98.9%) cases. In Klebsiella spp., Linozolid was sensitive in 48 (75%) cases, while Colistin showed resistance 57 (89%) cases.
In Gram-positive bacteria, Enterococcus faecalis displayed the highest sensitivity to Amikacin and Meropenem, with all 103 cases (100%). Conversely, eight antibiotics-Amoxicillin, Ceftriaxone, Cefuroxime, Ciprofloxacin, Clindamycin, Cefipime, Fluconazole, and Sulphamethoxazole exhibited the highest resistance, with all 103 cases (100%).
For Staphylococcus aureus, Amikacin displayed notable sensitivity in 78 cases (96.3%), while Colistin exhibited resistance in 67 cases (87.7%). Among Staphylococcus spp., 67 cases (87%) were sensitive to Amikacin, but 55 cases (71.4%) demonstrated resistance to Clindamycin.
This study revealed that gram-negative bacteria causing pus infections exhibited a higher MDR rate at 70.3%, in contrast to gram-positive bacteria at 29.9%. The overall MDR rate of gram-negative bacteria in our study was slightly higher than research studies conducted in Ethiopia [20, 27] several years ago. This difference may be attributed to variations in the study population, as previous studies focused exclusively on hospitalized inpatients, where higher MDR strains are expected.
Among the predominant isolated gram-negative bacteria, E. coli (98.52%), Pseudomonas spp. (92.66%), and Proteus spp. (91.58%) displayed the highest MDR percentages, while Klebsiella spp. (87.5%) exhibited the lowest. These MDR rates for gram-negative bacteria in our study exceeded the rates reported in previous studies [20, 27]. On the other hand, gram-positive bacteria, particularly Enterococcus faecalis (92.23%) and Staphylococcus aureus (95.06%), exhibited high MDR percentages.
This study provided a comprehensive overview of the current scenario of AMR and MDR isolates, highlighting their proportion rates across different age groups, including both males and females. It allowed us to differentiate the AMR and MDR patterns of both Gram-negative and Gram-positive bacterial isolates separately. However, it is essential to acknowledge certain limitations in this study. Given that the research was conducted in specific districts, it may not fully represent the overall MDR profile in Bangladesh. To address this, a nationwide experimental study is anticipated in the near future to assess the MDR scenario across the entire country.
CONCLUSION
The study revealed that different microorganisms like Pseudomonas spp., Enterococcus faecalis, Proteus spp., E. coli, Staphylococcus aureus, Staphylococcus spp., and Klebsiella spp. were the most prevalent bacteria among the infected pus samples. Both Gram-positive and Gram-negative bacterial isolates showed significant levels of antibiotic resistance. Without knowing the nature of these antibiotic resistance completely, it is onerous for medical practitioner to prescribe appropriate medicine and reduce the cost of therapy. Therefore, the study may help the physician to prescribe the proper antibiotics that would be more effective for pus patients to reduce the severity of infection.
ACKNOWLEDGEMENTS
We would like to acknowledge the Department of Biochemistry and Molecular Biology at Mawlana Bhashani Science and Technology University and the Lab Zone, and Hormone Center of Tangail for their unconditional support. This work was partially supported by grants of Research Cell, MBSTU from University Grants Commission of Bangladesh and from Ministry of Science and Technology, Bangladesh Grant No: 12601-120005400-3631199.
AUTHOR CONTRIBUTIONS
MJI conceptualized this study. HA, FAN, GB, and MK drafted the manuscript. MJI, MKMU, RAJ, AN, and SRS critically reviewed the manuscript. FAN collected the information and HA did the data analysis. All authors approved the final version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Tarana MN, Fardows J, et al. Bacteriological profile of wound swab and their antimicrobial susceptibility pattern in shaheed suhrawardy medical college, dhaka. Journal of Shaheed Suhrawardy Medical College. 2019;11:65-8.
- [2]Balasubramanian B, Benit N, et al. Carbapenemases producing klebsiella pneumoniae from the pus of hospitalized patients: In-vitro antibiotic properties of streptomyces against multidrug resistant infectious bacteria. Journal of Infection and Public Health. 2021;14:892-7.
- [3]Huda N, Yusuf MA, et al. Antimicrobial sensitivity pattern of bacteria isolated from pus sample collected from a private diagnostic laboratory in rangpur district of bangladesh. Bangladesh Journal of Infectious Diseases. 2021;8:64.
- [4]Khanam RA, Islam MR, et al. Bacteriological profiles of pus with antimicrobial sensitivity pattern at a teaching hospital in dhaka city. Bangladesh J. 2018;5.
- [5]Ohalete C, Obi R, et al. Bacteriology of different wound infection and their antimicrobial susceptibility patterns in imo state nigeria. World J Pharm Pharm Sci. 2012;1:1155-72.
- [6]Morgan DJ, Okeke IN, et al. Non-prescription antimicrobial use worldwide: A systematic review. The Lancet infectious diseases. 2011;11:692-701.
- [7]Gibson MK, Forsberg KJ, et al. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. The ISME journal. 2015;9:207-16.
- [8]Sharpe A, Jackson A. Stomaching: A new concept in bacteriological sample preparation. Applied Microbiology. 1972;24:175-8.
- [9]Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in eskape pathogens. BioMed research international. 2016;2016.
- [10]Akram M, Shahid M, et al. Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in jnmc hospital aligarh, india. Annals of clinical microbiology and antimicrobials. 2007;6:1-7.
- [11]Alam MM, Islam MN, et al. Prevalence of multidrug resistance bacterial isolates from infected wound patients in dhaka, bangladesh: A cross-sectional study. International Journal of Surgery Open. 2021;28:56-62.
- [12]Nobel F, Akter S, et al. Prevalence of multidrug resistance patterns of escherichia coli from suspected urinary tract infection in mymensingh city, bangladesh. Journal of Advanced Biotechnology and Experimental Therapy. 2021;4:256-64.
- [13]Talukder M, Islam MS, et al. Detection of multidrug resistant salmonella spp. From healthy and diseased broilers having potential public health significance. J Adv Biotechnol Exp Ther. 2021;4:248-55.
- [14]Zuhora FT, Hosen MA, et al. Molecular characterization of multidrug-resistant bacteria isolated from the external and internal parts of the housefly.
- [15]Alam MM, Islam M, et al. Antimicrobial resistance crisis and combating approaches. Journal of Medicine. 2019;20:38.
- [16]Organization WH. Antimicrobial resistance: Global report on surveillance: World Health Organization; 2014.
- [17]Safain KS, Bhuyan GS, et al. Situation of antibiotic resistance in bangladesh and its association with resistance genes for horizontal transfer. BioRxiv. 2020:2020.04. 06.027391.
- [18]Muluye D, Wondimeneh Y, et al. Bacterial isolates and their antibiotic susceptibility patterns among patients with pus and/or wound discharge at gondar university hospital. BMC research notes. 2014;7:1-5.
- [19]Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at dessie health research laboratory, ethiopia. Asian Pacific journal of tropical biomedicine. 2014;4:164-8.
- [20]Godebo G, Kibru G, et al. Multidrug-resistant bacterial isolates in infected wounds at jimma university specialized hospital, ethiopia. Annals of clinical microbiology and antimicrobials. 2013;12:1-7.
- [21]Ahmed I, Rabbi MB, et al. Antibiotic resistance in bangladesh: A systematic review. International Journal of Infectious Diseases. 2019;80:54-61.
- [22]Misha G, Chelkeba L, et al. Bacterial profile and antimicrobial susceptibility patterns of isolates among patients diagnosed with surgical site infection at a tertiary teaching hospital in ethiopia: A prospective cohort study. Annals of Clinical Microbiology and Antimicrobials. 2021;20:1-10.
- [23]Shakir A, Abate D, et al. Magnitude of surgical site infections, bacterial etiologies, associated factors and antimicrobial susceptibility patterns of isolates among post-operative patients in harari region public hospitals, harar, eastern ethiopia. Infection and Drug Resistance. 2021:4629-39.
- [24]Nobel FA, Islam S, et al. Isolation of multidrug resistance bacteria from the patients with wound infection and their antibiotics susceptibility patterns: A cross-sectional study. Annals of Medicine and Surgery. 2022;84:104895.
- [25]Trojan R, Razdan L, et al. Antibiotic susceptibility patterns of bacterial isolates from pus samples in a tertiary care hospital of punjab, india. International journal of microbiology. 2016;2016.
- [26]Jesmin H, Ahasan HN, et al. Antimicrobial resistanceamong intensive care unit patients in a tertiary care hospital of bangladesh. Bangladesh Journal of Medicine. 2021;32:5-11.
- [27]Yishak A, Biruk LW. Microbial susceptibility of bacteria isolated from open fracture wounds presenting to the err of black-lion hospital, Addis Ababa University, Ethiopia. African Journal of Microbiology Research. 2009;3(12):939-51.