Influence of the high mobility group A1 genetic polymorphism on indices of metabolic syndrome and insulin resistance in the Iraqi population: Case-control study
Abstract
The high mobility group A1 gene (HMGA1) rs139876191 variant has been related to metabolic syndrome and type 2 diabetes, but data are lacking in Middle Eastern populations. The study aimed to assess whether the HMGA1 rs139876191 variant is associated with metabolic syndrome risk and whether this variant predicts the risk of insulin resistance. This case-control study was carried out at single center in Kirkuk city/ Iraq from February to August 2022. Polymorphisms in HMGA1 and genotyping were identified by Sanger sequencing of genomic DNA obtained from 91 Iraqi participants (61 patients with metabolic syndrome and 30 control). Lipid profile, serum (glucose and insulin), glycated hemoglobin, blood pressure, body mass index, and waist circumference were also measured. The high prevalence of the del/del genotype of rs139876191 was found. Minor allele frequency of rs139876191 was 0.16 in both metabolic syndrome and the control group. A non-significant difference in genotyping was identified between total metabolic syndrome and the control group. The del/ins variant was associated with significantly higher waist circumference, triglycerides (TG), low-density lipoprotein (LDL), very low-density lipoprotein (VLDL), and glycated hemoglobin (HbA1c) (P=0.03, 0.041, 0.007, 0.034, and 0.001, respectively), and significantly lower high-density lipoprotein (HDL) (p=0.000). Linear regression analysis showed no significant effect of the variant (del/ins) on developing insulin resistance. Thus, rs139876191 polymorphism with del/ins genotype in the HMGA1 gene was not associated with metabolic syndrome risk but it was associated with indices of metabolic syndrome including waist circumference, TG, HDL, LDL, VLDL, and HbA1c. Besides, this variant did not predict the risk of insulin resistance.
INTRODUCTION
Metabolic syndrome (MetS) is a group of metabolic dysregulate including insulin resistance, hypertension, atherogenic dyslipidaemia, and central obesity [1]. The national cholesterol education program (NCEP) adult treatment panel III (ATP III) criteria define metabolic syndrome as the presence of any three of the following five traits: abdominal obesity, defined as a waist circumference ≥102 cm (40 in) in men and ≥88 cm (35 in) in females, serum triglycerides ≥150 mg/dL (1.7 mmol/L) or drug treatment for elevated triglycerides, serum HDL-cholesterol <40 mg/dL (1 mmol/L) in males and <50 mg/dL (1.3 mmol/L) in females or drug treatment for low HDL cholesterol, and blood pressure ≥130/85 mmHg or drug treatment for elevated blood pressure [2]. Individuals with MetS are at high risk for severe complications, like type 2 diabetes mellitus (T2DM) and heart diseases [3]. The prevalence of MetS varies between ethnic groups, diagnostic standards, and gender, indicating that there are related genetic factors underlying the etiology of the disease [4]. The high prevalence of MetS is particularly prevalent in certain ethnic groups, especially in the Middle East [5] and Asia [6]. A diet with high energy-rich foods has long been implicated as a factor in the etiology of MetS [7]. Genetic factors may affect MetS itself or each component of MetS [8]. Estimates of MetS heritability vary between 10% and 30% [4], indicating that MetS is partially hereditary. Diabetes is a group of metabolic disorders described by high blood sugar due to defects in insulin release, insulin action, or both [9, 10]. People with diabetes have lately been considered one of the critical triggers of heart failure, ischemic heart disease, and cardiovascular diseases [11, 12]. Insulin resistance is responsible for many diseases with a remarkable social impact, such as T2DM and MetS, which include the main risk causes for cardiovascular disease (like hypertension, hyperglycemia, hyperlipidemia, and obesity) [13]. The homeostatic model assessment of insulin resistance (HOMA-IR) is a mathematical model that employs the levels of fasting glucose and insulin to determine insulin resistance. High mobility group A1 (HMGA1) positively regulates the activity of the Insulin Receptor (INSR) promoter, which binds to the transcription starting point of INSR leading to positive regulation of INSR expression and insulin signal transduction [14]. The rs139876191 variant (also called IVS5-13insC) is present at position 13 of HMGA1 exon 6; Low-frequency insertion polymorphism of rs139876191 has been identified and associated with insulin resistance and T2DM among individuals of white European ancestry [15] and Chinese populations [16]. The effect of HMGA1 was studied in two populations (Italy and Turkey), both affected by MetS, results indicate that the variant HMGA1 rs139876191 was significantly associated with MetS, where this occurred irrespectively of the presence of T2DM, confirming the assumption that IVS5-13insC could independently relate with additional traits associated to insulin resistance [17]. The rs139876191 variant also associated with a certain metabolic syndrome-associated trait (i.e., high fasting plasma glucose, low HDL, reduced insulin sensitivity, high body mass index) was seen among individuals of European and Hispanic American ancestry [18]. This study was conducted to assess whether the HMGA1 rs139876191 variant is associated with metabolic syndrome risk and its related components and whether this variant predicts the risk of an increase in insulin resistance in a sample of the Iraqi population.
MATERIALS AND METHODS
Study population
This case-control study was done at Kirkuk city/ Iraq, internal medicine clinic under the supervision of an internal medicine specialist from February until August 2022. One hundred subjects were selected to participate in this study. Only 91 subjects finished this study successfully. These subjects were enrolled into the following groups: Two patient groups: 61 patients were divided into two main groups. The First Group contained 31 metabolic syndrome patients with T2DM. The Second group contained 30 metabolic syndrome patients without diabetes. The control group included 30 apparently healthy subjects who had no components of Mets criteria.
Patients with metabolic syndrome and apparently healthy subjects over 30 years old of either sex were accepted to contribute to the study. Metabolic syndrome was well-defined by National Cholesterol Education Program (NCEP) Adult Treatment Panel III (ATP III) guidelines [2].
Patients with type 1 diabetes or any type of malignancy, pregnant and lactating women, autoimmune diseases, and patients with Inborn Errors of metabolism were excluded from this study.
The study protocol was approved with the number (RECAUBCP4102021B) on the 4th of October 2021 by the Ethical Committee of the University of Baghdad-College of Pharmacy. Verbal informed consent was obtained from the participants.
Demographical, anthropometric, and biochemical evaluation
Data was collected using a researcher-made questionnaire which consisted of individual factors including age, gender, marital status, educational level, living place, smoking history, and the presence of diabetes mellitus, dyslipidemia, and hypertension. Body mass index (BMI) was calculated as weight divided by height (kg/m2). Systolic/diastolic blood pressure (SDBP) was measured with an MDF® desk mercury sphygmomanometer. Blood samples were collected following 12 h overnight fasting. Fasting serum glucose was measured by the enzymatic colorimetric method using a glucose oxidize test. Glycated hemoglobin was determined by the latex-enhanced immunoassay method. Serum total cholesterol, triglyceride, HDL, and LDL were determined by enzymatic colorimetric methods using commercial kits provided by (GIESSES®DIAGNOSTICS, Italy). Very low-density lipoprotein cholesterol was calculated as about one-fifth of triglyceride levels [19]. The sandwich electrochemiluminescence immunoassay (ECLIA) method was used to determine serum insulin by using the commercial kit provided by (Elecsys insulin, Cobas®, Germany).
Fasting serum glucose and insulin were obtained in those with MetS patients without DM and the control group who were not taking glucose tolerance-affecting drugs (such as nonselective beta-blocker, thiazide diuretics, and glucocorticoids, etc.) to calculate HOMA-IR, beta cells function and insulin sensitivity which calculated with HOMA2 calculator using http://www.dtu.ox.ac.uk/homacalculator/index.php.
Sample collection
Blood collected from patients and control after at least 12 hours of fasting took up to 5 ml of venous blood, and 2 ml was added to ethylene di-amine tetra acetic acid (EDTA) tube for detection of single nucleotide polymorphism (SNP) for HMGA1.
Primers
The DNA sequence of the HMGA1 gene was obtained from the NCBI GenBank database. Polymerase chain reaction (PCR) primers made by Primer Premier 3 software (Table 1), with 58 to 62°C melting temperatures, a primer length of 18 to 23 nucleotides, and PCR amplicon lengths of 800 to 1000 base pairs.
Table 1. The sequences of the primers.
Variant analysis
Genomic DNA was isolated from blood samples according to the ReliaPrep™ Blood gDNA Miniprep System protocol, Promega. A QuantusTM fluorometer was used to measure the concentration of extracted DNA and to determine sample quality for subsequent applications. Polymerase chain reaction amplification was performed by using PCR Express Thermal Cycler, BioRad, USA. Agarose gel electrophoresis was performed to check the presence or absence of amplification. Polymerase chain reaction products were sent to Macrogen Corporation, South Korea for Sanger sequencing using an automated DNA sequencer.
Statistical analysis
The statistical analysis was conducted using Statistical Package for the Social Sciences (SPSS) software for Windows version 26.0 (IBM Corp., Armonk, NY, U.S.). Continuous variables were expressed in mean ± standard deviation (SD) for normally distributed data and median (interquartile range) for skewed distributed data. Allele and genotypes were presented in number and frequency. The Shapiro-Wilk test was used to test the normality of the continuous variables. The Independent T-test was used for normally distributed data and the Mann-Whitney U test was used for not normally distributed data to determine a significant difference in demographic characteristics between the groups. The chi-square test or Fisher exact test was used to test group differences in proportions. Odds ratios (OR) and 95% confidence intervals were additionally calculated. Each quantitative trait was tested for normality and log-transformed when necessary. Univariate linear regression analysis was used to compare quantitative biochemical traits between wild and carriers after adjusting for covariates. A p-value of <0.05 was reflected as statistically significant.
RESULTS
Demographic, anthropometric, and biochemical characteristics of the study groups
The demographic, anthropometric, and biochemical characteristics of all participants involved in this study are shown in Table 2. Waist circumference (WC), BMI, total cholesterol, triglycerides, LDL, VLDL, SDBP, HbA1c, and fasting blood glucose (FBG) were higher in metabolic syndrome patients than in the control group as illustrated in Table 3.
Table 2. Demographic, anthropometric, clinical, and biochemical features of all participants.
Table 3. Anthropometric, and biochemical features of patients with metabolic syndrome.
Sanger sequences data
The extracted DNA concentration in all samples was found to be in a range of 20- 35ng/μl detected by using Quantus fluorometer TM. Analysis of rs139876191 SNP of HMGA1 gene using Sanger sequencing (Figure 1). Six “C” peaks reveal of C insertion homozygous allele. Five “C” peaks were revealing of a C Deletion homozygous allele. The presence of the “CCCCC” and “CCCCCC” peaks was revealing of del/ins heterozygous allele.
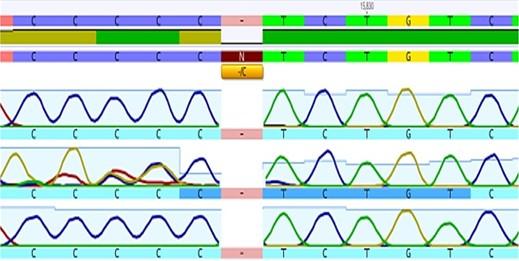
Prevalence of genotypes polymorphism
The high prevalence of the del/del genotype of rs139876191 was identified in the Iraqi population enrolled in the current study. Minor allele frequency of rs139876191 was (MAF=0.16) in both metabolic syndrome patients and the control group with the higher frequency seen in those who had diabetes with (MAF=0.23) as illustrated in Table 4.
Table 4. Distribution of HMGA1 genetic polymorphism (rs139876191) in Iraqi populations.
Comparison among different groups in genotyping
The non-significant difference in genotyping and allele carriage frequencies of the HMGA1 gene between total metabolic syndrome patients and the control group was found as illustrated in Table 5.
The odd ratio for developing metabolic syndrome with diabetes was 1.6 times (not significant) for the del/ins genotype (CI=0.58-4.64) compared to the del/del genotype of rs139876191 SNP as illustrated in Table 6.
Table 5. Comparison of genotypes and alleles frequency between total metabolic syndrome patients and control group.
Table 6. Comparison of the genotypes and alleles frequency between metabolic syndrome patients with diabetes and control group.
Effects of HMGA1 gene variant on indices of metabolic syndrome
There were significant differences in WC, TG, HDL, LDL, VLDL, and HbA1c (P=0.03, 0.041, 0.000, 0.007, 0.034, and 0.001 respectively) between the wild and the carrier genotype. Carriers of a del/ins variant were associated with low HDL and higher waist circumference, TG, LDL, VLDL, and HbA1c levels compared to those with the wild genotype of rs139876191 SNP as shown in Table 7.
Table 7. Effects of the rs139876191 variant on indices of metabolic syndrome.
Effects of the HMGA1 gene polymorphism on insulin
Fasting serum glucose and insulin were calculated in a subgroup of Iraqi subjects (30 MetS and 30 healthy) who were not taking glucose tolerance-impacting drugs and for whom glucose and insulin levels were obtainable. Linear regression analysis, adjusted for age, sex, and BMI displayed no significant effect of the variant (del/ins) on developing insulin resistance as illustrated in Table 8.
Table 8. Effects of the rs139876191 variant on insulin resistance.
DISCUSSION
The high mobility group A1 plays an essential role in glycemic balance as a structural transcription factor. Although this study is intended to reproduce the association between metabolic syndrome and HMGA1, it is one of a kind to try to establish an association of people of non-European origin. The current study thus emerges as a new finding in a Middle Eastern/Iraqi population in Kirkuk province. Interestingly, the HMGA1 SNPs were rare in the European population [13]. However, in this study, about 20 (32.8%) of metabolic syndrome patients were heterozygous with a minor allele frequency (MAF) of rs139876191 (MAF=0.16), about 10(33.3%) of the control group were heterozygous with a minor allele frequency of rs139876191 (MAF=0.16) and about 14(45.2%)of metabolic syndrome with T2DM were heterozygous with a minor allele frequency of rs139876191 (MAF=0.23) compared with other population studies where the MAF in metabolic syndrome (Italian 4.42% and Turkish 5.8%) and diabetes patients were (Hispanic American 21.4%, Italian 2.1%, American 3.1%, French 3.3%, Turkish 4.8%, Chinese 8.6%) [15, 16, 17, 18]. This could reflect the genetic heterogeneity among diverse ethnicities. This variant was existing in 32.8 % of MetS compared with 33% of controls which is inconsistent with another study that found that this variant was existing in 9-11 % of Italian and Turkish MetS compared with 4-7.5% of controls [17].
Metabolic syndrome has been associated with T2DM [20]. The rs139876191 variant considered one of the most significant genetic risk factors for T2DM yet descriptively, with odds were similar or even higher to (OR = 1.63) [21] which is similar to this study with (OR= 1.6) risk for developing T2DM. The precise biological mechanisms underlying the correlation between the HMGA1 gene and T2DM risk stay unclear [22].
The choice of a suitable control group is of significant importance and is one of the hardest features of case-control studies on insulin resistance and T2DM. The main problem is that many of these individuals will eventually develop T2DM and associated diseases, including hypertension, dyslipidemia, and cardiovascular disease if there is a personal or family history of insulin resistance. Therefore, without full testing, it is hard to rule out people with insulin resistance in a control group, so, control groups can contain a significant number of people with insulin resistance who will lead to T2DM in the future. To decrease the number of insulin-resistant individuals in Iraq's control groups, in this study, individual interviews were conducted with 30 healthy individuals to ensure there is no personal or family history (as a minimum the first relative degree) of T2DM and associated conditions, including high blood pressure, dyslipidemia, and premature cardiovascular disease.
A number of recent genome-wide association studies (GWASs) of T2DM have been described, but no link was detected between the variant HMGA1 rs139876191 and the existence of T2DM and metabolic syndrome [22]. Because T2DM is a complex disease, it is hard to identify all genetic risk factors from several GWAS studies, although GWASs are powerful tools in the study of complex diseases [16].
In this study, the mutant homozygote (ins/ins) was not appeared because of its low-frequency variant besides the small sample size which was already been seen in the study of the Chinese population that involve three groups of participants and the group that contain only 96 participants recruited from individual undergo routine health examinations at Tongi hospital in Wuhan were ins/ins was not recorded compared with group involve large sample size where ins/ins genotype was detected [16].
The data of this study suggest that low-frequency variants, including IVS5-13insC, are not associated with metabolic syndrome or even diabetes risk in the Iraqi population consistent with the study of the French population which found no association between this variant and T2DM [23]. Furthermore, in this study, the (del/ins) variant had significant effects on indices of metabolic syndrome including waist circumference, TG, HDL, LDL, VLDL, and HbA1c, where HDL is a main factor of MetS, which is commonly regarded as an independent predictor of major heart problem [24] and no significant association was seen with further quantitative traits of the metabolic syndrome. While another study displayed that the IVS5-13insC variant was associated with low HDL, high FBG, and BMI [17]. Low HDL level is significant and consistent with previous study, this indicates that HMGA1 can affect genes complicated in cholesterol homeostasis [25]. There is no significant effect (del/ins) variant on developing insulin resistance which is consistent with study including the French population [23] and inconsistent with another study that shows that the IVS5-13insC variant plays a part in the pathogenesis of MetS and other diseases linked to insulin resistance [17].
This study was limited by its small sample size and focus on a single center in one city (Kirkuk city) Thus, caution must be applied when generalizing the outcomes of this study to the whole country. Moreover, the study was conducted during the COVID-19 era, and the overall number of patients included was limited due to repeated curfews that resulted in the loss of several patients as a result of their inability to attend the clinic. This study also encountered a practical problem, which is that the del/ins mutation did not appear clearly in some samples when making the sequence in the forward direction, so we had to perform the sequence in the reverse direction as well.
CONCLUSIONS
This study found that the rs139876191 polymorphism with del/ins genotype in the HMGA1 gene was not associated with metabolic syndrome risk, but it was associated with indices of metabolic syndrome including waist circumference, triglyceride, high-density lipoprotein, low-density lipoprotein, very low-density lipoprotein, and HbA1c. Besides, this variant did not predict the risk of an increase in insulin resistance in Iraqi populations.
ACKNOWLEDGEMENT
Our sincere thanks to all participants who wholeheartedly contributed to this study. This research received no external funding.
AUTHOR CONTRIBUTIONS
The work was designed, supervised, and performed by Mirna, Eman and Omar. The first draft of this manuscript was prepared by Mirna and Eman. Mirna analyzed the data and improved the overview of the manuscript. Collecting data was done by Mirna and Omar. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Fahed G, Aoun L, Bou Zerdan M, et al. Metabolic syndrome: Updates on pathophysiology and management in 2021. Int J Mol Sci. 2022;23(2):786.
- [2][Huang PL. A comprehensive definition for metabolic syndrome. Dis Model Mech. 2009;2(5-6):231-237.
- [3]Azzawi OF. Metabolic syndrome; comparing the results of three definition criteria in an Iraqi sample. Al-Kindy Col. Med. J. 2018;14(2):7-12.
- [4]Carson C, Lawson HA. Epigenetics of metabolic syndrome. Physiol Genomics. 2018;50(11):947-955.
- [5]Ansarimoghaddam A, Adineh HA, Zareban I, Iranpour S, HosseinZadeh A, Kh F. Prevalence of metabolic syndrome in Middle-East countries: Meta-analysis of cross-sectional studies. Diabetes Metab Syndr. 2018;12(2):195-201.
- [6]Grundy SM. Metabolic syndrome: a multiplex cardiovascular risk factor. J Clin Endocrinol Metab. 2007;92(2):399-404.
- [7]Velasquez MT. Altered Gut Microbiota: A link between diet and the metabolic syndrome. Metab Syndr Relat Disord. 2018;16(7):321-328.
- [8]Pokharel S, Acharya S, Chowdhury ATMM. Genes associated with metabolic syndrome and hyperuricemia: An overview. Clin Case Reports Rev. 2015;1(7):143–8.
- [9]Fakree NK, Ali SH: Effect of COX-2 Inhibitors Selectivity on Lipid Profile in Hyperlipidemic and Normolipidemic Type 2 Diabetics. Iraqi J. Pharm. Sci. 2009; 18(Suppl): 7–13.
- [10]Mikhael EM, Hassali MA, Hussain SA, Shawky N. Self-management knowledge and practice of type 2 diabetes mellitus patients in Baghdad, Iraq: a qualitative study. Diabetes Metab Syndr Obes. 2018;12:1- 17.
- [11]Obaidullah M, Chowdhury M, Islam S, Barman A, Hossain I, Matin M. Analysis of the risk of cardiovascular diseases among people with diabetes according to triglyceride level. Journal of Advanced Biotechnology and Experimental Therapeutics. 2021;4(2):210.
- [12]Mahmud M, Hasan M, Roy D, Ullah M, Basak B, Reza M, et al. Tumor necrosis factor-alpha 308G>A polymorphism cause coronary heart disease in type-2 diabetic patients of the northern region of Bangladesh. Journal of Advanced Biotechnology and Experimental Therapeutics. 2021;4(1):60.
- [13]Chiefari E, Foti DP, Sgarra R, et al. Transcriptional Regulation of Glucose Metabolism: The Emerging Role of the HMGA1 Chromatin Factor. Front Endocrinol (Lausanne). 2018;9:357.
- [14]Semple RK, Savage DB, Cochran EK, Gorden P, O'Rahilly S. Genetic syndromes of severe insulin resistance. Endocr Rev. 2011;32(4):498-514.
- [15]Chiefari E, Tanyolaç S, Paonessa F, et al. Functional variants of the HMGA1 gene and type 2 diabetes mellitus. JAMA. 2011;305(9):903-912
- [16]Liu L, Ding H, Wang HR, et al. Polymorphism of HMGA1 is associated with increased risk of type 2 diabetes among Chinese individuals. Diabetologia. 2012;55(6):1685-1688.
- [17]Chiefari E, Tanyolaç S, Iiritano S, et al. A polymorphism of HMGA1 is associated with increased risk of metabolic syndrome and related components. Sci Rep. 2013;3:1491.
- [18]Pullinger CR, Goldfine ID, Tanyolaç S, et al. Evidence that an HMGA1 gene variant associates with type 2 diabetes, body mass index, and high-density lipoprotein cholesterol in a Hispanic-American population. Metab Syndr Relat Disord. 2014;12(1):25-30.
- [19]Sahu S. Calculation of VLDL-cholesterol from triglycerides and total cholesterol levels. Biomedicine 2008;28(3)219-221.
- [20]Jabbar TL, Kasim AA. Association of retinol binding protein- 4 (RBP4) with glycemia, dyslipidemia, hypertension, and obesity in type 2 diabetic Iraqi patients. Iraqi J Pharm Sci. 2021;29(2):263–70.
- [21]Bianco A, Chiefari E, Nobile CG, Foti D, Pavia M, Brunetti A. The association between HMGA1 rs146052672 variant and type 2 diabetes: A transethnic meta-analysis. PLoS One. 2015;10(8):e0136077.
- [22]Liu Y, Zheng L, Kong H, Wang Q, Tian X. HMGA1 variant IVS5-13insC is associated with insulin resistance and type 2 diabetes: an updated meta-analysis. Afr Health Sci. 2018;18(4):865-872.
- [23]Marquez M, Huyvaert M, Perry JR, et al. Low-frequency variants in HMGA1 are not associated with type 2 diabetes risk. Diabetes. 2012;61(2):524-530
- [24]Barter P, Gotto AM, LaRosa JC, et al. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357(13):1301-1310.
- [25]Treff NR, Pouchnik D, Dement GA, Britt RL, Reeves R. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene. 2004;23(3):777-785.