Biofilm formation by the interaction of fungi (Candida tropicalis) with various bacteria
Abstract
Biofilms are composed of more than one species and are often called polymicrobial biofilms, so research is needed on the formation of polymicrobial biofilms, especially between fungi and bacteria. This study aimed to analyze the total biomass and metabolic activity of biofilms formed from the interaction of fungi (Candida tropicalis) with various bacteria, including Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, Pseudomonas aeruginosa, and Acinetobacter baumannii. Bacterial and fungal cultures were suspended in tryptic soy broth (TSB) medium, and biofilm was cultivated in two 96-well microplates for 48 hours at 37°C. The crystal violet assay was used to detect the total biomass biofilm, and the tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, or MTT) assay was used to obtain the metabolic activity of biofilm with the optical density (OD) value using an ELISA reader. The results of this study obtained significant data from both parameters. The highest average value was found in the C. tropicalis treatment group (2,412 ± 0.825 on the biomass test results; 1,525 ± 0.473 on the metabolic activity test results), while the lowest was seen in the S. aureus treatment group (0.512 ± 0.224 in the biomass test results; 0.978 ± 0.349 in the metabolic activity test results). In conclusion, it was confirmed that biofilm biomass could be generated strongly in each treatment in the total biomass parameter, while metabolic activity data suggested that each treatment could carry out cell proliferation.
INTRODUCTION
Fungal infections with bacteria, often known as polymicrobial infections have increased. Polymicrobial infection is an interaction caused by the formation of polymicrobial biofilms, especially between fungi and bacteria [1]. Polymicrobial biofilm is an interaction process of multispecies microbes to form a biofilm community [2]. Polymicrobial biofilm infections have a higher risk of death (about 70%) than infections caused by a single species of microbe (23%) [3]. Microbial biofilms are an important research area for two reasons. Firstly, understanding the form and existence of bacteria in multicellular communities is an intriguing basic science. Secondly, the formation of biofilms can cause problems in several fields, including medical and industrial. Additionally, biofilms can cause antimicrobial treatment resistance, decreased host response, and biocides resistance [4]. Several factors affect the type of interaction between fungi and bacteria depending on the species, strain, and environmental parameters such as temperature, pH, and glucose levels [1, 5]. Candida tropicalis is an opportunistic pathogen with a high infection rate. This microbe is one of the most common pathogens of the non-albicans candida (NAC) species and has the highest adhesion ability compared to other NAC species [6–8].
In polymicrobial communities, several mechanisms can occur, including antagonism mechanisms that drive competition for nutrients and cooperative mutualism synergism mechanisms that provide a mutual benefit under certain conditions [9, 10]. The communication mechanism between microorganisms is often associated with quorum sensing (QS), a signaling molecule that plays a role in coordinating the expression of virulence and microbial survival [5, 11]. Each cell is capable of responding to quorum-sensing molecules (QSMs) [12, 13]. QSMs also play an important role in biofilm development, morphogenesis, cell population restriction, infection emergence, and dissemination processes [11, 14].
Candida spp. is an example of the interaction between fungi and bacteria, since it interacts with the gram-positive bacteria Staphylococcus aureus, which can mutually increase the level of virulence and resistance in the host [1]. Fungal and bacterial interactions can occur in various infections, for example, Candida spp. with Pseudomonas aeruginosa, Acinetobacter baumannii, and methicillin-resistant S. aureus (MRSA). The interaction between Candida albicans and MRSA can coexist synergistically to form biofilms [3, 15].
Since polymicrobial biofilms cause severe infection, this study analyzes the effect of polymicrobial biofilms derived from interactions between the fungi C. tropicalis and the bacteria S. aureus, MRSA, P. aeruginosa, and A. baumanii on total biomass and metabolic activity of biofilm formation.
MATERIALS AND METHODS
Ethical approval
This research has received ethical approval with certificate reference number: 138/HRECC.FODM/III/2022 on March 31, 2022, by the Faculty of Dental Medicine Health Research Ethical Clearance Commission Universitas Airlangga.
Sample collection
The strains were obtained from the Microbiology Laboratory, Faculty of Medicine, Universitas Airlangga, Indonesia. Strains were stored at 4°C in saboraud dextrose agar (SDA) medium (Oxoid) for Candida tropicalis strain and tryptic soy agar (TSA, OXOID) medium for bacterial strains. Each strain is a clinical isolate from Dr. Soetomo General Hospital, Surabaya, Indonesia. This research was conducted at the Microbiology Laboratory, Faculty of Medicine, Universitas Airlangga, Indonesia from March to May 2022.
Preparation of microbial suspension
Prior to inoculation, all strains were re-cultured from the initial culture to their respective growth medium: SDA medium for Candida tropicalis, and TSA medium for bacterial strains. Then all that had been re-cultured were incubated aerobically at 37°C (Binder Model BD 400) for 24 h [16]. After incubation, the suspension of each strain was formed by inoculating each strain into 5 ml of tryptic soy broth (TSB) medium (OXOID, CM0129B), and incubated aerobically at 37°C for 24 h. Microbes from the suspension were vortexed and compared for turbidity with a McFarland 0.5 solution (equivalent to 1×107 CFU/ml) [17].
Biofilm cultivation
The microbial suspension was used in the biofilm cultivation process on two different 96-well microplates (NEST, flat bottom TC sterile, Nest Biotechnology Co., Ltd.). The first microplate was used for the CV assay parameters and the second microplate was used for the MTT assay parameters. The biofilm cultivation step was carried out by inoculating each microbial suspension into 9 treatment wells: one well for the control group (containing only 150 µL of TSB medium), five wells given a single-species suspension of 150 µL each (C. tropicalis, S. aureus, MRSA, P. aeruginosa, and A. baumanii), and four wells were inoculated with 75 µL of mixed-species suspension to obtain a volume of 150 µL in one well. Each treatment was repeated 6 times, so there were 60 filled wells. After the process, the microplate was closed, covered with plastic wrap, and incubated at 37 degrees Celsius for 48 hours without shaking [18, 19].
Crystal violet assay
In this method, the total biomass was obtained by removing the non-adherent cells from each well and rinsing the adherent cells using phosphate-buffered saline (PBS, pH 7.2) three times. Subsequently, the microplate was air-dried for approximately 30 minutes. A total of 150 µL of CV solution (0.4% (w/v), Ceristain C.I 42555) was poured into each well, allowed to stand for 15 minutes, then the microplate was rinsed under running water and air-dried at room temperature. After drying the microplate, each treatment well was fixed by adding 150 µL of 95% ethanol (Onemed, Jayamas medical industry, Indonesia). Following this procedure, the microplate was ready for reading the absorbance value with an ELISA reader (Thermo multiscan Go 1510, Thermo Fisher Scientific, Inc.) at 570 nm [16, 19, 20].
MTT assay
This method is a colorimetric test that provides a value for metabolic activity due to the reduction of the MTT tetrazolium salt. The first step of the method was to remove non-adherent cells in each well and rinsed with PBS (pH 7.2) twice. Furthermore, 15 µL of MTT solution (3-(4,5-dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Code M6494, invitrogenTM) which had been dissolved with PBS was poured into each well including control wells. Then the microplates were incubated at room temperature 37°C for 4 hours in the dark. After incubation, 150 µL of dimethyl sulfoxide solution (DMSO, Vivatis code PC0906, Vivantis Technologies Sdn Bhd-Malaysia) was added to the wells to remove the remaining medium and dissolve the MTT formazan crystals. Furthermore, the color change from yellow to purplish in each well was observed on the microplate. After this process, the microplate was ready to read the absorbance value using an ELISA reader (BioTek 50TM, MERCK) at 540 nm [21–23].
Data analysis for crystal violet assay
The data obtained from the crystal violet assay was analyzed using the microtiter plate assay method by calculating the cut-off OD (ODc) value. In this method the biomass value was obtained and grouped based on its ability to produce biofilms with the following formula format as described [19,24]:
Data analysis for MTT assay
The data obtained from this method is the average value of OD in each well which was used to determine the level of metabolic activity which is characterized by the presence of cell proliferation, with the following formula as described [22,23],
Statistical analysis
In this study, each microplate underwent 10 treatments with 6 repetitions. Each result obtained is presented as mean ± standard deviation. The results obtained from the CV assay and MTT assay in the form of OD values were analyzed for differences using one-way ANOVA with Tukey’s post hoc follow-up test with SPSS version 16.0 software application with 95% confidence degree (P-value <0.05 is considered to be significantly different).
RESULTS
Effect of biofilm formation on total biomass as measured by CV assay
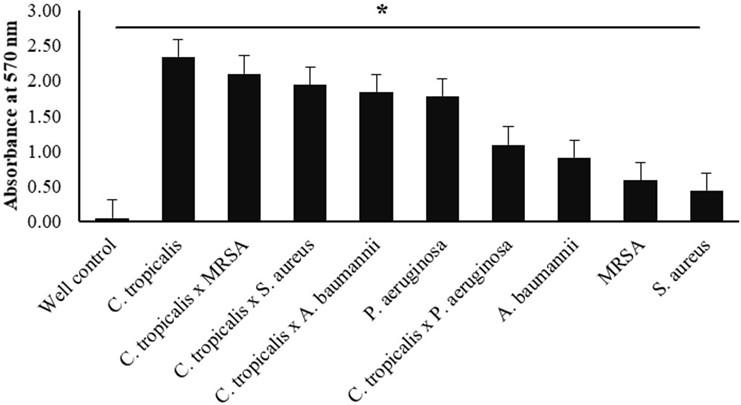
In this study, cultivation was carried out on single-species and fungal-bacterial biofilms (Candida tropicalis, Staphylococcus aureus, MRSA, Pseudomonas aeruginosa, and Acinetobacter baumanii). The biofilms were grown in 96-well microplates and incubated for 48 h. Crystal violet staining was performed, and the absorbance value was measured three times with an ELISA reader at 570 nm. The results showed that biofilm biomass was formed in both treatments (single-species and mixed-species), except in control wells (TSB medium only), with the difference between the mean OD isolate and the mean ODc being 0.072 (data not shown). This indicates that each OD isolate value is four times greater than the ODc value (0.072). Based on the previously described formula, each treatment is included in the high biofilm-forming (HBF) category.
Each treatment data set was presented as mean ± SD from the highest to the lowest value, respectively. Single-species treatment of Candida tropicalis had the highest total biomass value (2,412 ± 0.825) and S. aureus had the lowest value (0.512 ± 0.224) treatment (Table 1).
Table 1. Biofilm formation for each treatment was measured by CV staining and MTT method.
Based on the results described in Table 1, the graph (Figure 1) explains the differences in the OD-treatment values from the highest in C. tropicalis to the lowest in S. aureus. The X-axis represents the type of treatment species, while the Y-axis shows the OD-treatment value at a wavelength of 570 nm. The error bar indicates the standard deviation value. The one-way ANOVA statistical test showed a significant (P-value <0.05) difference between the single-species and mixed-species biofilm treatments.
Effect of biofilm formation on metabolic activity by MTT assay
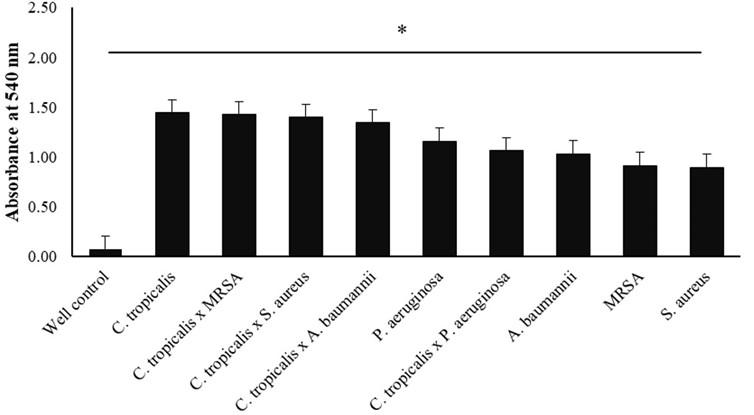
In this method, single-species and mixed-species biofilm cultivation were carried out on Candida tropicalis, Staphylococcus aureus, MRSA, Pseudomonas aeruginosa, and Acinetobacter baumanii strains. The biofilm was grown in a 96-well microplate for 48 hours before the tetrazolium salt method test (3-(4,5-dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium bromide). The presence of metabolic activity in the biofilm is indicated by the reduction or discoloration of the tetrazolium salt from yellow to blue formazan. These results were obtained by measuring the OD value three times with an ELISA reader at 540 nm, then calculating the difference between the average value of OD in each treatment with the average value of OD-control, which shows 0.079 (table 1).
The results are presented as mean ± SD for each treatment, following the previously described formula regarding grouping the results of cell proliferation activity. Thus, this study found the category of increased cell proliferation (ICP) in the following treatments, C. tropicalis (1,525 ± 0.473), C. tropicalis x MRSA (1,508 ± 0.380), C. tropicalis x S. aureus (1,481 ± 0.364), and C. tropicalis x A. baumannii (1,423 ± 0.304). In addition, this study also found the results of cell proliferation which were included in the normal cell proliferation (NCP) category in the treatment of P. aeruginosa (1,239 ± 0.500), C. tropicalis x P. aeruginosa (1.144 ± 0.095), A. baumannii (1.114 ± 0.313), MRSA (0.995 ± 0.165), and S. aureus (0.995 ± 0.165) (Table 1).
As described in table 1, the graph (Figure 2) shows the highest OD value (treatment with ICP category for C. tropicalis) to the lowest OD value (treatment with NCP category for S. aureus). The X-axis indicates the treatment of species, while the Y-axis shows the OD value at a wavelength of 540 nm. The error bar indicates the standard deviation value (Figure 2). The one-way ANOVA statistical test showed a significant (P-value <0.05) difference between the single-species and mixed-species.
DISCUSSION
In this study, the formation of in-vitro biofilms showed significant results. These findings indicated that each strain was able to form biofilms. The crystal violet dye binds to negatively charged surface molecules from living and dead cells, and exopolysaccharides can be used to calculate total biomass [25, 26]. MTT assay method reduces yellow-colored tetrazolium salt to blue-colored formazan due to the activity of mitochondria or enzymes in plasma cells such as oxidoreductase, dehydrogenase, and oxidase. So in this method cell, proliferation and viability were calculated using this method [27, 28].
This study demonstrates that the single-species C. tropicalis treatment yielded the highest absorbance value for both parameters. (Table 1). This is because the C. tropicalis strain is a yeast capable of producing hyphae for attachment [29]. According to another study, C. tropicalis exhibited higher adhesion than other Candida species [30]. Hyphae in Candida play a role in the preparation of exopolymer substances and blastopores, this is what causes C. tropicalis biofilms to form strongly [31]. The C. tropicalis strain formed strong biofilm [32].
Biofilms have complex structures that can adhere to both biotic and abiotic surfaces [29]. In vitro biofilm formation has three important steps namely, attachment, colonization, and cell proliferation [33]. Another study showed that there are five important steps in the formation of fungal and bacterial biofilms, namely, adsorption, adhesion, and formation of microcolonies along with the matrix. The maturation process is characterized by metabolic activity and the emergence of quorum sensing (QS) molecules, and the dispersion process [1]. Biofilm formation is based on the concept of microbial community formation, which is mediated by a cascade of molecular mechanisms and gene expression. The quorum sensing (QS) molecular signal contained in each microbe plays an important role in communication between microbes to form a population. [12,34].
One of the factors that influence the biofilm is cell attachment to the substrate. In C. tropicalis, the adhesion molecule and the regulation of the BCR1 and RBT5 genes play an important role in attachment [29, 35]. S. aureus strains can form biofilms on the host or other surfaces. This attachment is mediated by recognizing adhesive matrix molecules (MSCRAMMs), regulated by the ica locus gene and hemB mutation [36]. Furthermore, this strain can produce extracellular matrix factor and polysaccharide intercellular adhesin (PIA), which is regulated by the Ica gene and is involved in cell multiplication [37].
In this study, the absorbance values of the CV assay and MTT assay of mixed-species C. tropicalis with MRSA were higher than that of C. tropicalis with S. aureus. This was because the MRSA strain was more virulent in terms of mortality and morbidity than the S. aureus strain with MSSA [38]. MRSA is encoded by the mecA gene, which binds to the PBP 2A protein and confers resistance to methicillin and beta-lactam drugs [39]. This study supports previous findings that mixed-species biofilms had lower absorbance values than fungal biofilms because bacteria can prevent Candida spp. development [40].
Findings of this revealed that the mixed-species treatment of C. tropicalis with gram-positive bacteria MRSA and S. aureus had a higher absorbance value than the interaction of C. tropicalis with gram-negative bacteria P. aeruginosa and A. baumannii. The findings of this investigation differ from prior studies on Candida albicans with gram-negative (E. coli and P. aeruginosa) and gram-positive (S. aureus) bacteria. It showed that gram-negative bacteria were able to reduce C. albicans biofilm compared to gram-positive bacteria [41]. P. aeruginosa has the virulence factor flagellin which plays a role in motility and has type III secretion [42]. A. baumannii has the polysaccharide capsule gene K1 [43]. Antagonism arises in mixed-species P. aeruginosa because this strain can kill hyphae and biofilms of C. tropicalis, which are triggered by phenazine compounds that damage cell wall integrity [44, 45]. However, this study is consistent with prior in vitro investigations on dual biofilms that show P. aeruginosa can limit the production of non-albicans Candida (NAC) biofilms such as C. tropicalis. This is because both microbes can release quorum sensing molecules and the presence of N- acyl homoserine lactone (AHL) in gram-negative bacteria. While in vitro conditions can decrease AHL in P. aeruginosa, inhibiting Candida spp. biofilms [46, 47].
In this experiment, single-species data on biomass or metabolic activity of bacteria exhibited a lower absorbance value than mixed species data [3]. In A. baumanni and MRSA, the lowest absorbance value was found in S. aureus. This is because S. aureus colonizes more slowly and forms biofilms on abiotic surfaces or under monoculture conditions. The colonization of this strain increases when in mixed biofilm conditions because the microcolonies of this strain can attach to Candida spp. hyphae. This indicated that C. tropicalis was able to facilitate the growth of S. aureus [48]. Based on the preceding description, this study found that there is a difference in total biomass value and metabolic activity between biofilm development from single-species and mixed-species treatments.
CONCLUSION
Findings of the current study indicated that the aqueous leaf extracts of X. americana and P. capensis confer cardiocurative activities in rats induced with myocardial infarction. However, further studies are recommended to test and compare cardiocurative treatment effects of various phytochemicals identified in the studied extracts.
ACKNOWLEDGEMENT
We acknowledge to Institute of Tropical Disease, Universitas Airlangga, Surabaya, Indonesia, and Microbiology Laboratory, Research Center Faculty of Dental Medicine, Universitas Airlangga, Surabaya, Indonesia.
AUTHOR CONTRIBUTIONS
DWA did the research proceed and conceived the manuscript, ADWW and RJS planned the idea for this research, S provided direction for data analysis, and WT and MRW helped to revise this manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Costa-orlandi CB, Sardi JCO, Pitangui NS, Oliveira HC De, Marcelino MY, Braz JD, et al. Fungal biofilms and polymicrobial diseases. Journal of Fungi 2017;3:1–24.
- [2]Gabrilska RA, Rumbaugh KP. Biofilm models of polymicrobial infection. Future Microbiol 2015;10:1997–2015.
- [3]Zago CE, Silva S, Sanitá PV, Barbugli PA, Dias MI, Lordello VB, et al. Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and resistant Staphylococcus aureus (MRSA). PLoS One 2015;10:1–15.
- [4]Harmsen M, Yang L, Pamp J, Tolker-nielsen Tim. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol 2010;59:2532–68.
- [5]Roy PK, Ha AJW, Mizan MFR, Hossain MI, Ashrafudoulla M, Toushik SH, et al. Effects of environmental conditions (temperature, pH, and glucose) on biofilm formation of Salmonella enterica serotype Kentucky and virulence gene expression. Poultry Science 2021;100:101209.
- [6]Muadcheingka T, Tantivitayakul P. Distribution of Candida albicans and non-albicans Candida species in oral candidiasis patients : Correlation between cell surface hydrophobicity and biofilm forming activities. Arch Oral Biol 2015;60:894–901.
- [7]Hutnick MA, Pokorski JK. Polymeric interventions for microbial infections : A review. Mol Pharmaceutics 2018;15:2910–2921.
- [8]Silva-dias A, Miranda IM, Branco J, Monteiro-soares M, Pina-vaz C, Rodrigues AG. Adhesion , biofilm formation , cell surface hydrophobicity , and antifungal planktonic susceptibility : relationship among Candida spp. Front Microbiol 2015;6:1–8.
- [9]Peters BM, Jabra-rizk MA, May GAO, Costerton W, Shirtliff ME. Polymicrobial interactions : Impact on pathogenesis and human disease. Clin Microbiol Rev 2012;25:193–213.
- [10]Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. Journal of Microbiology 2014;52:188–199.
- [11]Wongsuk T, Pumeesat P, Luplertlop N. Fungal quorum sensing molecules : Role in fungal morphogenesis and pathogenicity. J Basic Microbiol 2016;56:440–447.
- [12]Albuquerque P, Casadevall A. Quorum sensing in fungi – a review. Med Mycol 2012;50:337–345.
- [13]Cássia J De, Sardi O, Souza N De, Rodríguez-arellanes G, Lucia M, Fusco-almeida AM, et al. Highlights in pathogenic fungal biofilms. Rev Iberoam Micol 2014;31:22–29.
- [14]Ramage G, Saville SP, Thomas DP, López-Ribot JL. Candida biofilms: An update. Eukaryot Cell 2005;4:633–638.
- [15]Dixon EF, Hall RA. Noisy neighbourhoods: quorum sensing in fungal-polymicrobial infections. Cell Microbiol 2015;17:1431–1441.
- [16]Stepanović S, Vuković D, Hola V, Bonaventura G Di, Djukić S, Ćirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007;115:891–899.
- [17]Leonhard M, Zatorska B, Moser D, Schneider-Stickler B. Growth media for mixed multispecies oropharyngeal biofilm compositions on silicone. Biomed Res Int 2019;2019:1–8.
- [18]Tan Y, Leonhard M, Schneider-Stickler B. Evaluation of culture conditions for mixed biofilm formation with clinically isolated non-albicans Candida species and Staphylococcus epidermidis on silicone. Microb Pathog 2017;112:215–220.
- [19]Corte L, Casagrande Pierantoni D, Tascini C, Roscini L, Cardinali G. Biofilm specific activity: A measure to quantify microbial biofilm. Microorganisms 2019;7:1–14.
- [20]Capote-Bonato F, Sakita KM, de Oliveira AG, Bonfim-Mendonça P de S, Crivellenti LZ, Negri M, et al. In vitro interaction of Candida tropicalis biofilm formed on catheter with human cells. Microb Pathog 2018;125:177–182.
- [21]Gulati M, Lohse MB, Ennis CL, Gonzalez RE, Perry AM, Bapat P, et al. In vitro culturing and screening of candida albicans biofilms. Curr Protoc Microbiol 2018;50:e60.
- [22]Plumb JA. Cell Sensitivity Assays: The MTT Assay. In: Langdon S., editor. Cancer Cell Cult. Methods Mol. Med., vol. 88, Humana Press; 2004, p. 165–169.
- [23]van Meerloo J, Kaspers GJL, Cloos J. Cell Sensitivity Assays: The MTT Assay. In: Cree IA, editor. Cancer Cell Cult. Methods Protoc. Methods Mol. Biol., vol. 731. Second, Humana Press; 2011, p. 237–245.
- [24]Kırmusaoğlu S. The methods for detection of biofilm and screening antibiofilm activity of agents. antimicrob. Antibiot. Resist. Antibiofilm Strateg. Act. Methods, IntechOpen; 2019, p. 1–17.
- [25]Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods 2007;72:157–165.
- [26]Costa ACBP, Pereira CA, Freire F, Junqueira JC, Jorge AOC. Methods for obtaining reliable and reproducible results in studies of Candida biofilms formed in vitro. Mycoses 2013;56:614–622.
- [27]Wady AF, Machado AL, Foggi CC, Zamperini CA, Zucolotto V, Moffa EB, et al. Effect of a silver nanoparticles solution on Staphylococcus aureus and Candida spp. J Nanomaterials 2014;2014:1–7. https://doi.org/10.1155/2014/545279.
- [28]Präbst K, Engelhardt H, Ringgeler S, Hübner H. Basic Colorimetric Proliferation Assays: MTT, WST, and Resazurin. In: Gilbert DF, Friedrich O, editors. Cell Viability Assays. Methods Mol. Biol., vol. 1601, Humana Press, New York; 2017, p. 1–17.
- [29]Zuza-Alves DL, Silva-Rocha WP, Chaves GM. An update on Candida tropicalis based on basic and clinical approaches. Front Microbiol 2017;8:1–25.
- [30]Kaur R, Dhakad MS, Goyal R, Kumar R. Emergence of non-albicans Candida species and antifungal resistance in intensive care unit patients. Asian Pac J Trop Biomed 2016;6:455–460.
- [31]Pires RH, Montanari LB, Martins CHG, Zaia JE, Almeida AMF, Matsumoto MT, et al. Anticandidal efficacy of cinnamon oil against planktonic and biofilm cultures of Candida parapsilosis and Candida orthopsilosis. Mycopathologia 2011;172:453–64.
- [32]Silva S, Rodrigues CF, Araújo D, Rodrigues ME, Henriques M. Candida species biofilms’ antifungal resistance. J Fungi 2017;3:1-17.
- [33]Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, et al. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J Dent Res 2001;80:903–908.
- [34]Araújo D, Henriques M, Silva S. Portrait of candida species biofilm regulatory network genes. Trends Microbiol 2016;25:62–75.
- [35]Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol 2006;8:1382–91.
- [36]Gordon RJ, Lowy FD. Pathogenesis of methicillin‐resistant Staphylococcus aureus infection. Clin Infect Dis 2008;46:S350–9.
- [37]Götz F. Staphylococcus and biofilms. Mol Microbiol 2002;43:1367–78.
- [38]Gastmeier P, Sohr D, Geffers C, Behnke M, Daschner F, Rüden H. Mortality risk factors with nosocomial Staphylococcus aureus infections in intensive care units: Results from the German nosocomial infection surveillance system (KISS). Infection 2005;33:50–5.
- [39]Grema HA, Geidam YA, Gadzama GB, Ameh JA, Suleiman A. Methicillin resistant staphylococcus aureus (mrsa): a review. Adv Anim Vet Sci 2015;3:79–98.
- [40]Hager CL, Isham N, Schrom KP, Chandra J, McCormick T, Miyagi M, et al. Effects of a novel probiotic combination on pathogenic bacterial-fungal polymicrobial biofilms. American society for microbiol 2019;10:1–11.
- [41]Park SJ, Han K, Park JY, Choi SJ, Lee K. Influence of bacterial presence on biofilm formation of candida albicans. Yonsei Med J 2014;55:449–58.
- [42]Boisvert A-A, Cheng MP, Sheppard DC, Nguyen D. Microbial biofilms in pulmonary and critical care diseases. Ann Am Thorac Soc 2016;13:1615–23.
- [43]Amala Reena Aa, Subramaniyan A, Kanungo R. Biofilm formation as a virulence factor of Acinetobacter baumannii: An emerging pathogen in critical care units. J Curr Res Sci Med 2017;3:74.
- [44]Hogan DA, Kolter R. Pseudomonas – candida interactions: An ecological role for virulence factors. Science 2002;296:2229–32.
- [45]Morales DK, Jacobs NJ, Rajamani S, Krishnamurthy M, Cubillos-Ruiz JR, Hogan DA. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol 2010;78:1379–92.
- [46]Fourie, Pohl. Beyond Antagonism: The interaction between candida species and Pseudomonas aeruginosa. J Fungi 2019;5:34.
- [47]Navarro-Arias MJ, Hernández-Chávez MJ, Garcia-Carnero LC, Amezcua-Hernández DG, Lozoya-Pérez NE, Estrada-Mata E, et al. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect Drug Resist 2019;12:783–94.
- [48]Harriott MM, Noverr MC. Importance of Candida–bacterial polymicrobial biofilms in disease. Trends Microbiol 2011;19:557–563.