Next generation sequencing for the formalin-fixed paraffin-embedded samples of ovarian cancer in Vietnam
Abstract
Ovarian cancer is one of the most common causes of mortality among women, and the prevalence of ovarian cancer increases. Early diagnosis of this disease via genetic variant testing is one potential strategy for enhancing treatment and disease outcome. Our aim was to establish a standard procedure of next generation sequencing (NGS) for the formalin-fixed paraffin-embedded (FFPE) forms of ovarian tumor tissue to detect genetic mutation in our laboratory. Here, we used the FFPE samples of ovarian tumor tissues from Vietnamese patients to detect pathogenic variants in BRCA1/BRCA2 via the NGS. DNA was extracted using the QIAamp DNA FFPE Tissue Kit, and then its quality was assessed by the BioDrop and Qubit. The BRCAaccuTestTM PLUS kit and Illumina MiSeqDx instrument were used for both library preparation and sequencing. All samples had passed the A260/280 ratio cut-off for DNA purity and the requirement of DNA concentration. Excepted for the 1st time, the percentage of ≥ Q30 was more than 80%, while the density was approximately 1,200 K/mm2, while the phasing and prephasing (%) metrics were satisfied to be less than 0.1%. Five pathogenic variants in BRCA1/BRCA2, including both single nucleotide polymorphisms and indels were successfully detected using NgeneAnalySysTM software. In conclusion, DNA extraction from the FFPE sample was qualified for sequencing and the sequencing results met all the required metrics for variant analysis.
INTRODUCTION
Among malignant tumors in women, ovarian cancer (OC) is the eighth most common cancer cause of death in women worldwide [1] with the survival rate of OC being less than 45% [2]. It is also the leading cause of death of all gynecologic malignancies [3]. The lifetime risk of this type of cancer is around 1.3% in women [3]. According to GLOBOCAN 2020, 3000 women were diagnosed with this disease in 2020 [2]. In the USA, the estimate of OC was approximately 21,410 cases and 13,770 deaths in 2021 [1]. In Vietnam, a developing country with a population being nearly 100 million people, the age-standardized incidence rate of this disease is one of the lowest [2]. Around 2.4 cases per 100,000 population are diagnosed per year [2].
If OC is detected early, at stage I or II, the 5-year survival rates are 90% and 70%, respectively [3]. Nevertheless, most cases are diagnosed at stage III or IV, which reduces 5-year survival rates to under 30% [3]. The reason for this status of late diagnosis is that the symptoms of OC are not clear when it is at its early stages, but later patients can show various symptoms related to appetite, digestion, and abdominal pain [1]. Therefore, early cancer detection and especially screening trials for individuals with increased risk of OC are of absolute importance. The greatest risk factors of OC are family history and other genetic syndrome [1]. The cumulative risk of OC was 49% for breast cancer 1 (BRCA1) gene mutations, 21% for breast cancer 2 (BRCA2) gene mutations by the age of 80, and 11-15% by the age of 70 for women with Lynch syndrome mutations [3]. As a result, various platforms have been developed and clinically applied to detect gene mutation in OC patients for diagnostic and therapeutic purposes.
With the advancement of new technology, Next Generation Sequencing (NGS) is playing an increasingly important role in cancer research due to its unique advantages including test sensitivity, speed at a considerably low cost, and the ability to sequence all mutation types for hundreds to thousands of genes [4]. NGS examinations into numerous cancer subgroups such as ovarian, breast, prostate, and pancreatic cancer have shown novel cancer genes and their mutational profiles [4]. Genetic testing for some familial malignancy genes including BRCA1 and BRCA2 is recommended for individuals considered to be at high risk due to their family health status and clinical history [4]. In addition, NGS will easily replace other sequencing for the diagnosis of mutations in therapeutically significant genes in cancer tissues. For example, if the presence of the mutation is at a low level, NGS can still be efficient while Sanger sequencing may miss it [5]. The utilization of NGS at the beginning of the diagnosis process results in significant expense savings through the simultaneous sequencing of numerous specimens and targets, especially for subjects with malignancies similar to ovarian and breast cancer [4]. Therefore, every patient with epithelial OC is recommended to undergo testing for hereditary susceptibility genes [5].
In OC cases, formaldehyde-fixed paraffin-embedded (FFPE) samples are a valuable source of molecular information. Therefore, we conducted this study to describe the methods and application of NGS for FFPE samples of OC in our laboratory in Vietnam.
MATERIALS AND METHODS
Ethics approval
The research was approved by the Institute of Genome Research Institutional Review Board according to the decision number: 02-2022/NCHG-HĐĐĐ on March 09, 2022.
DNA extraction from FFPE samples
Genomic DNA of was extracted from 5 sections of 10 μm thickness of macro-dissected ovarian tumor tissues which contain at least 30% tumor cells by using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). According to the manufacturer's protocol, DNA extraction was performed following six main steps such as 1) Removing paraffin: paraffin was dissolved and removed by toluene; 2) Lysis: removed paraffin samples were lysed under denaturing conditions with proteinase K at 56°C in 1-8 hours; 3) Heat: sample and protein K mix was incubated at 90°C for reversing formalin crosslinking 1 hours; 4) DNA Binding: DNA binds to the membrane of filter column and contaminants flow through; 5) Washing: residual contaminants were washed away by washing buffers (WBI and WBII); and 6) Elution: pure, concentrated DNA was eluted from the membrane using 50-100μl elution buffer. Extracted DNA was stored at -80°C for further experiments.
DNA quantification and quality analysis
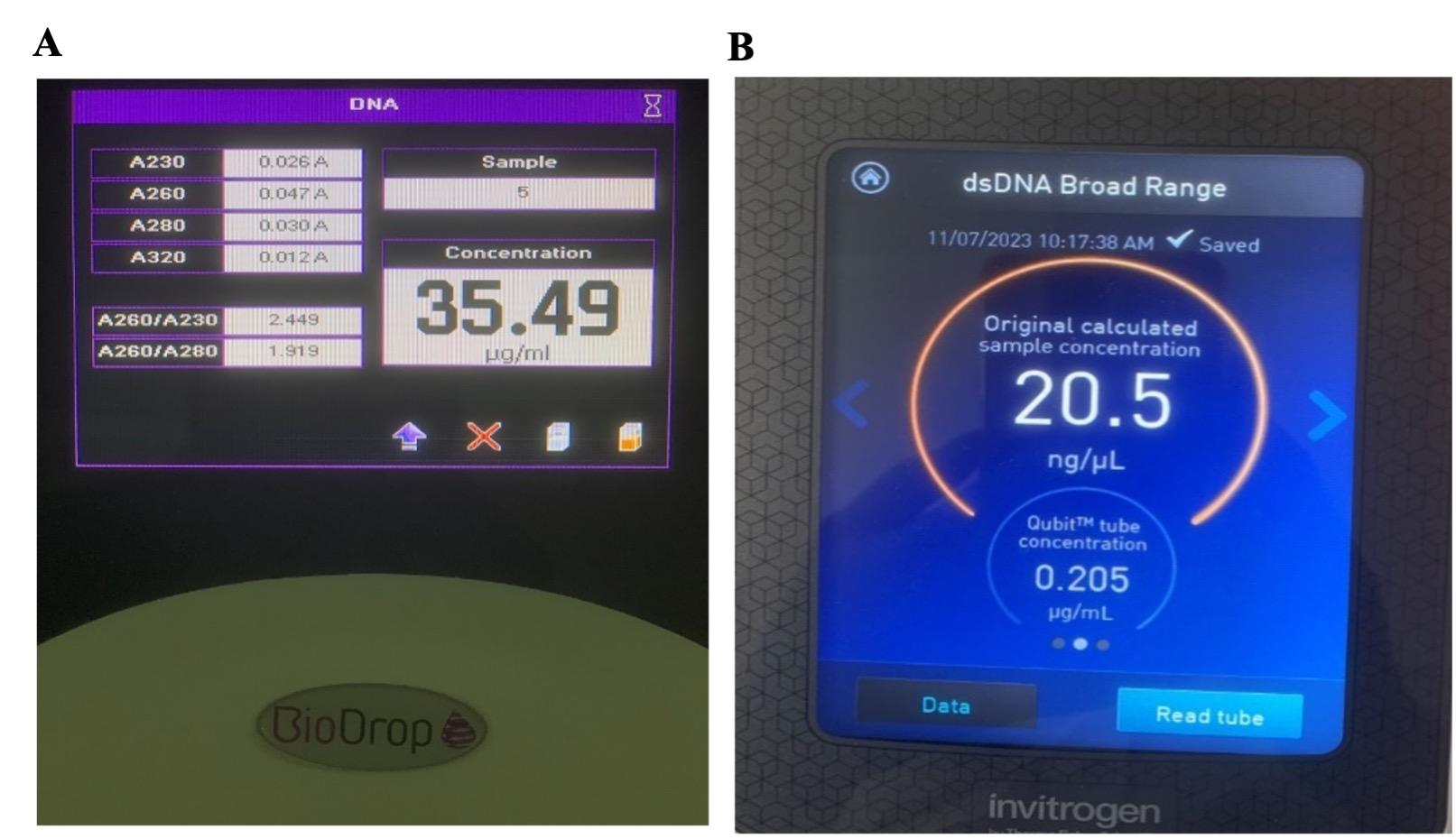
The quantitative process is measured by BioDrop UV-Visible spectrophotometer (Biochrom, Cambridge, The United Kingdom). In this stage, the concentration and purity of the DNA stock should be in the range of 20 – 200 ng/µL for the optimal result of further tests. The ratio of A260/280 is maintained at around 1.8 – 2.0 to ensure the purity of the DNA. The concentration of dsDNA is calculated by the following formulation: dsDNA concentration = 50 μg/mL × OD260 × dilution factor
Furthermore, Double-stranded DNA quantification was assessed by Invitrogen Qubit 4 Fluorometer (Thermo Fisher Scientific Inc, Massachusetts, USA). A Qubit dsDNA BR (broad range, 2 to 1000 ng) Assay Kit was used according to the manufacturer’s protocols; a sample volume of 2 μl was added to 198 μl of a Qubit working solution.
Next generation sequencing
In our study, the BRCAaccuTestTM PLUS kit (NGeneBio Co., Ltd, South Korea) and MiSeqDx instrument (Illumina Inc., USA) were used for analyzing BRCA1 and BRCA2 mutations in genomic DNA isolated FFPE tissue of OC patients.
According to the manufacturer, a total of 160 primer sets which cover all protein-coding exons and partial 5’-/3’-ends of BRCA1 and BRCA2 were designed to produce sequencing libraries with adapters and barcodes compatible with Illumina platform. The entire analyzed target size of BRCA1 and BRCA2 including all protein-coding regions, splicing regions, selected promoter, UTR, and intron regions were about 22.4 kb. The medium size of amplicons was 211 including primer sequences. BRCAaccuTestTM PLUS version NGB112V-012 is capable of running up to 11 somatic samples (+1 control DNA) simultaneously per run and MiSeq Reagent Micro v2 (300 cycles) was employed for sequencing.
The BRCA1 and BRCA2 genetic alteration analyzing procedure using BRCAaccuTestTM PLUS with NGS system consists of four main steps: sample preparation, library preparation, NGS data generation, and variants analysis. A total of 100ng of high-quality genomic DNA was used for two separate library preparation reactions.
Statistical analysis
Analysis software NgeneAnalySysTM (NGeneBio Co., Ltd, South Korea) was used for performing variants analysis. The reference materials and clinical specimens that carry BRCA point mutations, insertion, and deletion mutations were used to assess the sensitivity (Limit of Detection, LOD), specificity (interfering substance), precision (repeatability, reproducibility, and robustness), and accuracy (method comparison) of BRCAaccuTestTM PLUS. The criteria and suitability of the test results including mutation detection, heterozygosity mutation frequency, minimum coverage, and uniformity were established. The mutation detection rate and the heterozygote mutation frequency were defined as the positive percent agreement of each reference material and the ratio of the mutation (alternative) allele in the heterozygote, respectively. The minimum coverage was defined as the ratio of areas with a minimum number of 20 reads in the area subject to the BRCA1 and BRCA2 tests. The uniformity of sequencing was accessed to confirm that all tested areas had been evenly analyzed. The ratio of the areas over 20% of the average coverage was defined as uniformity. According to the manufacturer’s protocol, the recommended thresholds of the average coverage, minimum coverage, and uniformity for MiSeq performance are 1,500X, 100X, and 95%, respectively. Means were compared using Wilcoxon test. Statistics was performed by SPSS v22.0 (IBM, USA) with p < 0.05 as statistical significance.
RESULTS
DNA quantification and quality
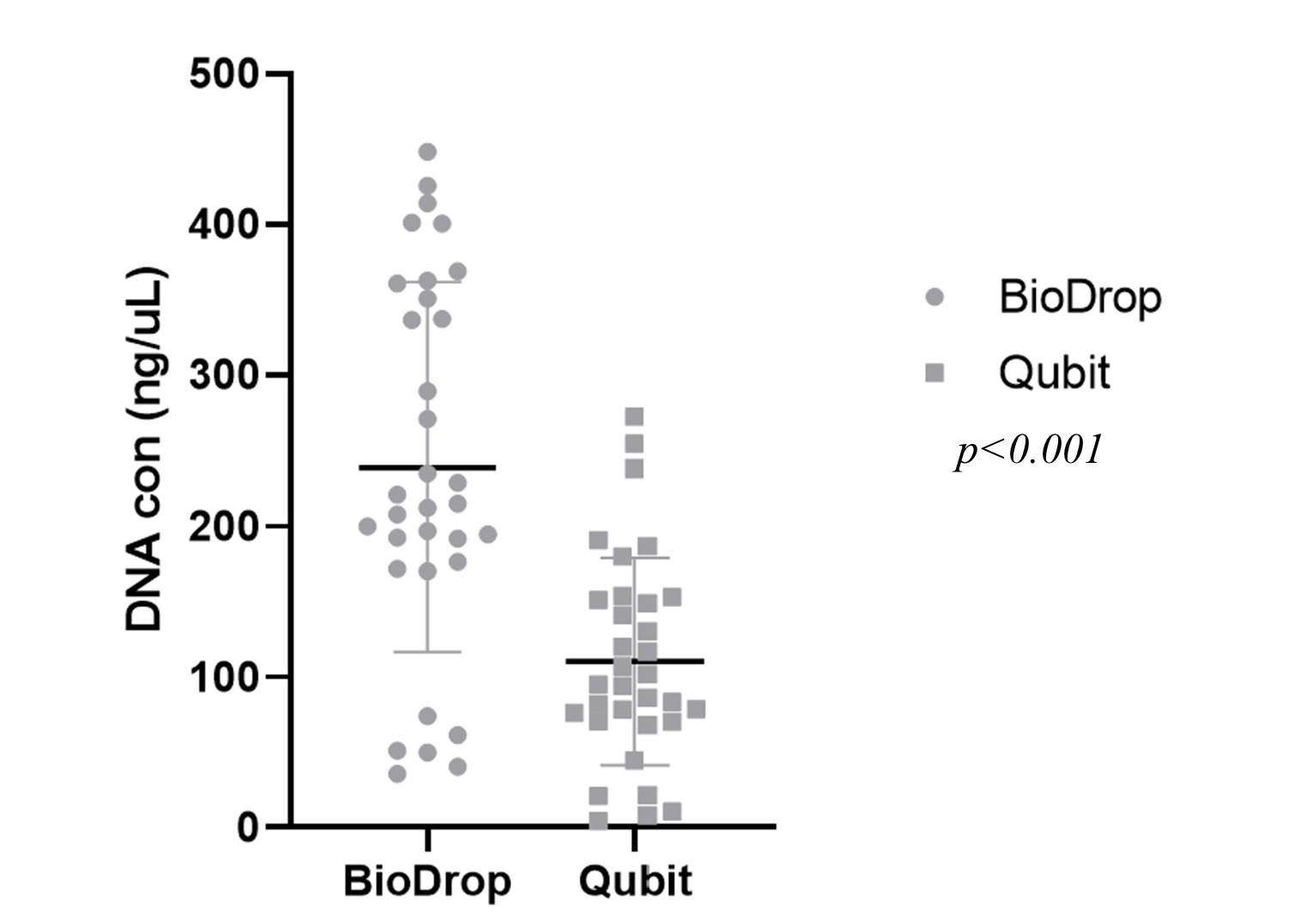
In this study, the quality of DNA extraction was evaluated based on the concentration of DNA (the threshold concentration was 20 – 200 ng/µL for later NGS) and the purity of DNA via an A260/280 ratio (with the threshold of 1.8 – 2.0). 33 samples were measured by both spectrophotometer and Fluorometer, and the results showed that all DNA extraction from samples passed the purity requirement (Table 1 and Figure 1). Only 2/33 samples (OC25 and OC16, accounted for 6.1%) had a DNA concentration higger than 200 ng/µL, thus they had to be diluted before sequencing by Illumina. BioDrop estimated a higher DNA concentration than Qubit (p <0.01) (Figure 2).
Table 1. DNA quantification and quality from 33 samples.
NGS quality control
NGS was successfully performed for 33 samples, which were divided into 3 runs, each run included 11 samples and 1 control DNA. Each run involved Read 1, 2, and 3. The NGS quality control was estimated by Sequencing Analysis Viewer (SAV) via several metrics, including yield total, % >= Q30, Density, Phas/Prephas (%), etc, (Table 2 and Figure 3). Except for Read 2 of Run 2, the percentage of bases with a quality score of more than 30 in all runs was higher than 80%, which is the recommended value for further analysis. Thus, although the number of reads in each run was around 6 million, the number of reads that passed the filter in Run 2 was 4.17 million, while this figure for Run 1 and 3 was higher (5.24 and 5.35, respectively). The cluster density of all runs was around 1,200 K/mm2, while the phasing and prephasing (%) metrics were satisfied to be less than 0.1% (Table 2) [6].
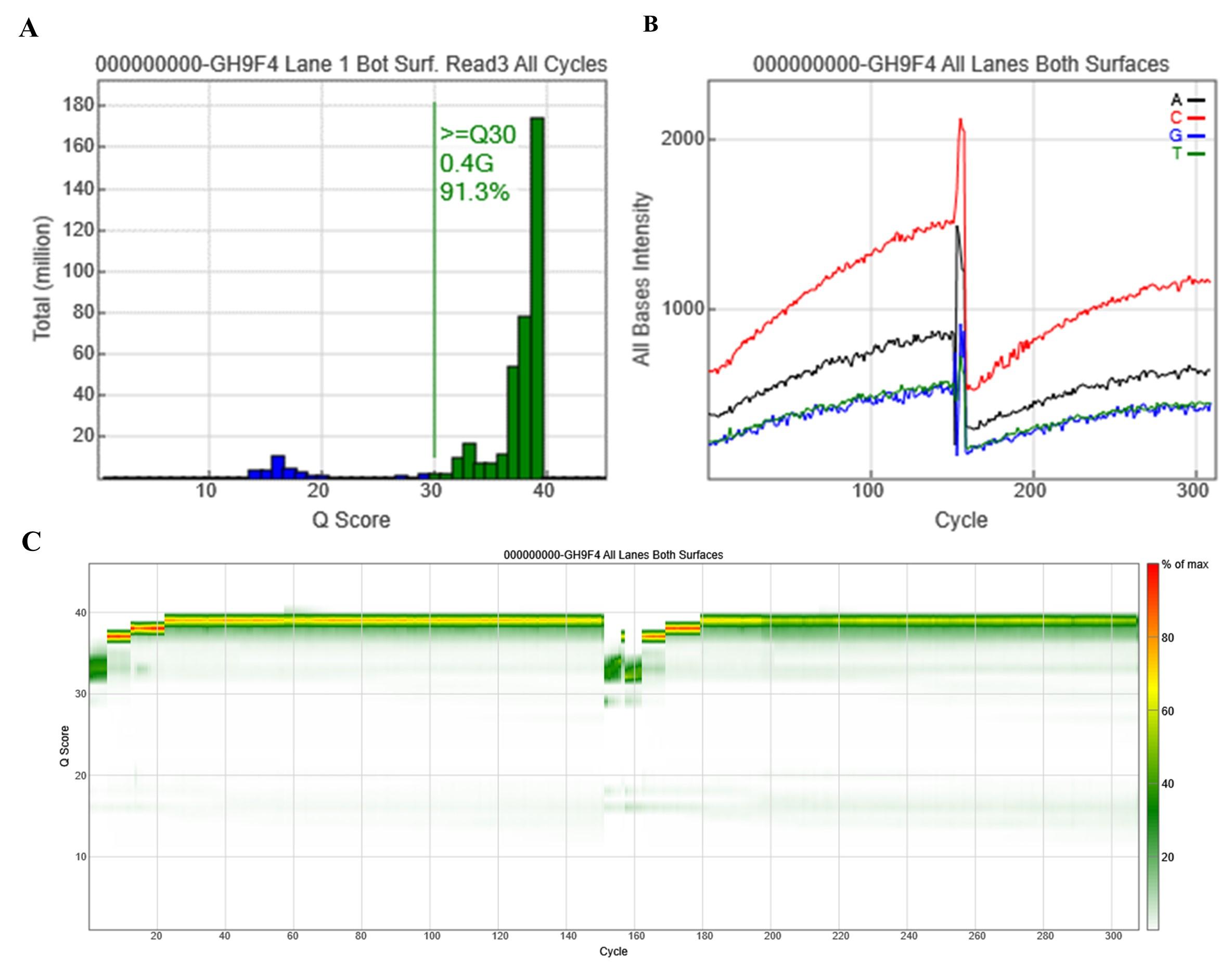
Table 2. Summary of NGS running metrics.
Variant detection
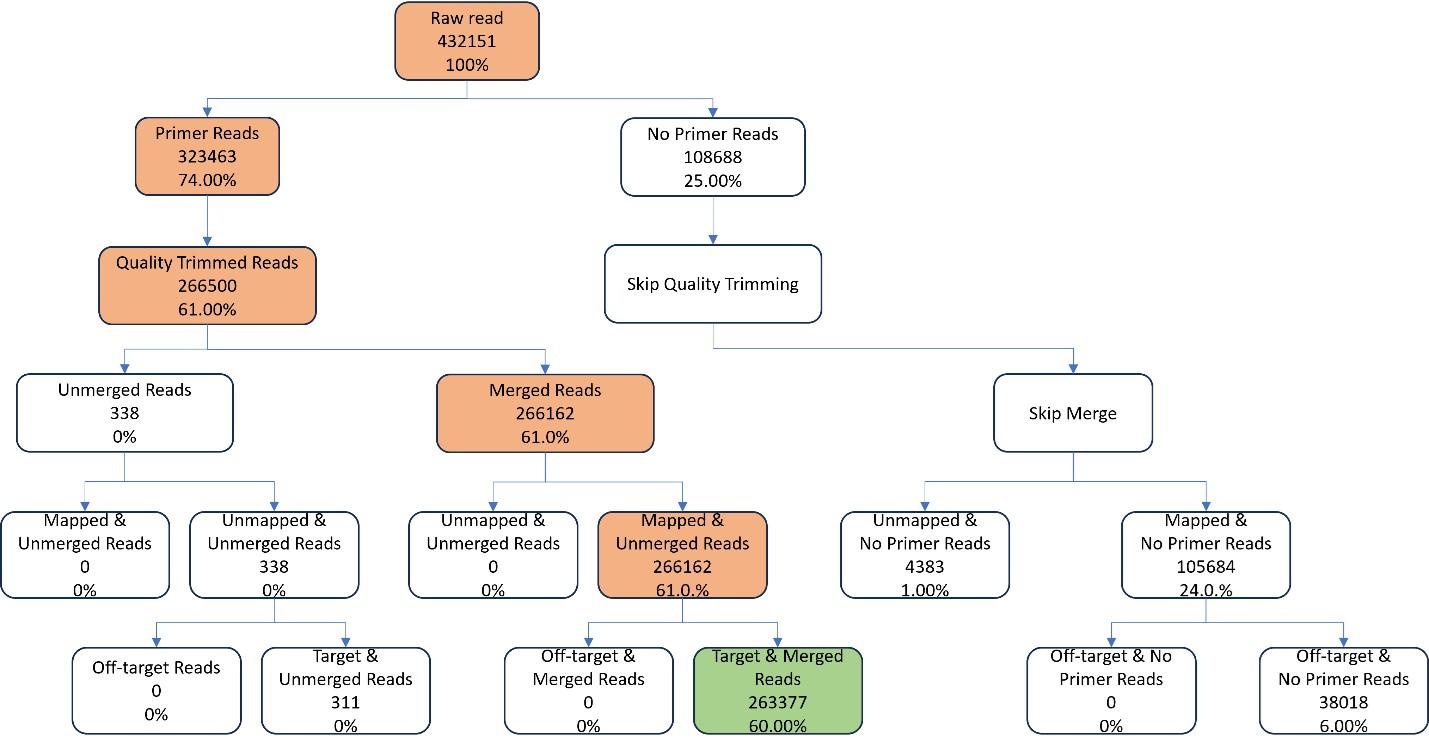
Sequencing data were then processed following several steps, including trimming, mapping, and merging. The total amplicon number was 160, while the average length of primers was 25bp and the average length of amplicon without primer was 158.756bp. The percentage of ROI region with coverage at least 20x was 100% and the percentage uniformity of coverage 0.2 was 100%. Among the paired-end raw reads of 432,151, only 60% was ready for variant calling (Figure 4).
Regarding variant calling, processed reads were aligned with the Human HG19 genome as a reference utilizing the BWA-MEM algorithm. The ubiquitous variant in Asian populations was excluded, including c.4563A>G, c.4563A>G, c.7397T>C. Several variants were detected in each sample, and related information of variants including ACMG pathogenicity classification, NT change, belonged gene, position, exon, dbSNP ID, etc, were provided (Table 3 and Figure 5).

Table 3. Variant calling of BRCA1/2 in OC48 samples.
DISCUSSION
Samples of common tumors in biobanks are frequently available, but most tumors with small sizes or very rare exist in FFPE pathology archives. However, there are challenges to FFPE sample availability including higher false-positive and false-negative rates of mutations than matched frozen tissues. This is due to DNA degradation and chemical contaminants caused by cross-links during formalin fixation. The number of functional copies of DNA is low compared to other types of samples and the deamination of DNA is the main cause of false positives [7]. Meanwhile, DNA extracted from FFPE samples is degraded after long-term storage. Guyard et al. observed a reduction in the maximal length of DNA fragments, indicating an increase in DNA fragmentation. The amount of remaining DNA obtained after 5.5 years only accounted for 11% of DNA collected at first extraction using qPCR and 47% using fluorimetry [8]. Moreover, a higher rate of mutations is detected in FFPE tissues compared to matched frozen tissues and it is difficult to recognize “artificial” mutations. Therefore, novel methodologies are constantly developed to reduce these errors substantially including pretreating with uracil-DNA-glycosylase. The performance of samples in NGS is also influenced by DNA extraction and library preparation approaches so assessing the quality of extracted DNA beforehand is helpful to select the suitable NGS method [9].
In this study, 33 FFPE samples of ovarian tumor tissue were successfully utilized to extract DNA for NGS sequencing. According to the BRCAaccuTestTM PLUS kit protocol, the required input DNA for the sequencing from the FFPE samples is higher than from the whole blood samples. Our BioDrop and Qubit results showed that both the requirement of the DNA concentration and purity were desired in all samples. To test the accuracy of BRCAaccuTestTM and NGeneAnalySys™ software, Kim et al. compared their analysis results to Sanger sequencing and indicated that the rate of concordance for both variants and wild-type locations was 100% in multiple samples. Regarding the performance metrics of BRCAaccuTestTM, 0.5 ng was the detection limit of single nucleotide variant, insertion, and deletion, while interfering substances were found to have no effect on analysis results and the reproducibility and repeatability showed 100% of precision, which was consistent with our results. Kim et al.’s study also indicated the stability of residual DNA samples and the rate of successful library preparation using BRCAaccuTestTM was 99.5% [10]. When comparing the results of sequencing running metrics, we found that the percentage of ≥ Q30, the cluster density, and phasing/prephasing were better than the results of several Miseq instruments [6].
Although most of the variants in BRCA1 and BRCA2 genes were classified as benign, 5 pathogenic and likely pathogenic variants were detected, including c.2865delC, c.1801_1808delCACAATTC, c.1673_1674delAA, c.1016delA and c.928C>T. 4/5 variants were deleted type, which then caused frameshift while the remaining led to stop-gained consequences. Only the c.2865delC variant was found in the BRCA2 gene, while the other 4 variants were found in the BRCA1 gene. This result was consistent with Kim et al.’s study while 13/15 pathogenic and likely pathogenic variants were found in the BRCA1 and only 3/15 variants were found in the BRCA2 gene. However, only the c.928C>T variant overlapped between the two studies although both participants were from Asia [10].
CONCLUSION
Despite the certain intrinsic obstacles of the FFPE sample, we successfully sequenced the BRCA1 and BRCA2 genes using NGS from FFPE tumor tissues of Vietnamese OC patients. The results showed that DNA extraction from the FFPE sample was qualified for sequencing and the sequencing results met all the required metrics for variant analysis. Thus, the FFPE sample is totally suitable for genetic testing. Variant calling performance detected several single nucleotide variants and indels in both BRCA1 and BRCA2 genes of OC patients. Although most of them were benign, 5 variants were indicated to be related to OC disease. However, further analysis is needed to clarify the relationship between BRCA1/BRCA2 variants and other demographic features and the risk of OC.
ACKNOWLEDGMENTS
This research is funded by Vietnam National University, Hanoi (VNU) under project "Researching on some clinical, non-clinical, and epidemiological characteristics, and genetic mutation and expression in Vietnamese patients with ovarian cancer" number: 776/QĐ-ĐHQGHN on March 26, 2021. We also would like to thank all members of the Center for Biomedicine and Community Health for data collection and helping to improve the manuscript.
AUTHOR CONTRIBUTIONS
DTC developed the ideas, designed the study, and conceptualized the manuscript; all authors collected the data, analyzed the data; DTC, QNN, NLB and TDV drafted the manuscripts; DTC and NLB revised and edited the manuscript; DTC supervised. All authors have read and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Chu D-T, Ngoc SMV, et al. The gene expression and mutations in ovarian cancer: Current findings and applications. Cham: Springer International Publishing. 2023; p. 1-19.
- [2]Le TN, Tran VK, et al. Brca1/2 mutations in vietnamese patients with hereditary breast and ovarian cancer syndrome. Genes (Basel). 2022;13.
- [3]Nebgen DR, Lu KH, et al. Novel approaches to ovarian cancer screening. Curr Oncol Rep. 2019;21:75.
- [4]Sabour L, Sabour M, et al. Clinical applications of next-generation sequencing in cancer diagnosis. Pathol Oncol Res. 2017;23:225-34.
- [5]Bonadio RC, Crespo JR, et al. Ovarian cancer risk assessment in the era of next-generation sequencing. Ann Transl Med. 2020;8:1704.
- [6]Kastanis GJ, Santana-Quintero LV, et al. In-depth comparative analysis of illumina® miseq run metrics: Development of a wet-lab quality assessment tool. Molecular Ecology Resources. 2019;19:377-87.
- [7]Gaffney EF, Riegman PH, et al. Factors that drive the increasing use of ffpe tissue in basic and translational cancer research. Biotech Histochem. 2018;93:373-86.
- [8]Guyard A, Boyez A, et al. DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch. 2017;471:491-500
- [9]Cazzato G, Caporusso C, et al. Formalin-fixed and paraffin-embedded samples for next generation sequencing: Problems and solutions. Genes (Basel). 2021;12.
- [10]Kim MJ, Song BJ, et al. Evaluation of a targeted next-generation sequencing assay for BRCA mutation screening in clinical samples. Lab Med Online 2021;11:283-9.