Developing Pseudomonas aeruginosa mutants with hyper-proteolytic activity through UV mutagenesis and characterization for optimized production
Abstract
Over 60% of the global productions of industrial enzymes are proteolytic enzymes in which about 35% are alkaline proteases. The current microbial sources are unable to reach industrial demands of alkaline protease which led scientists to search new sources with enhanced enzyme activity. Therefore, we applied UV irradiation to develop a Pseudomonas aeruginosa mutant as a new source of protease overproduction, followed by cultural and nutritional optimizations. The mutagenesis was carried out by exposing parent strains to UV radiation (30w, 2537 Å) at 25 ºC with a different time interval. The protease activity was estimated as relative protease activity and standard protease assay (OD660). Among all, mutant strain P. aeruginosa–M25 (PA-M25) exhibited 75.47% increased protease activity over the parent strain in submerged fermentation. It showed 612.84±2.50 U/ml of alkaline protease production compared to 349.26±2.57 U/ml by wild-type strain (significant at P≤0.005). Besides, the effects of nutritional factors on the protease production by PA-M25 were also studied. We found the optimal alkaline protease production in the medium (adjusted to pH 9.0) supplemented with 1% (w/v) glucose as carbon source, 0.5% (w/v) casein as nitrogen source and 2% (w/v) NaCl when incubated at 35 ºC for 48 h without agitation. We believe that the mutant PA-M25 could be a potential candidate to meet the growing protease demands. However, further assessments regarding the characterization of the protease enzyme, as well as the industrial fitness of the mutant, are warranted.
INTRODUCTION
Proteases (EC 3.4.21) are ubiquitous in biological systems and are well-known for their wide range of applications in food, detergent, and biomedical industries. Globally, proteases alone contribute ~65% market share among all industrial enzymes [1, 2] about 40% of which are microbial proteases [3]. Microbial proteases can catalyze the hydrolysis of proteins to release amino acids and serve many industrial applications. For instance, alkaline proteases mostly used in detergent formulations, which constitute 30% of the world’s total enzyme production, due to their stability and high activity at the higher pH range [4]. Laundry detergents usually maintain a pH range between 9-12, therefore, alkaline proteases are necessary for the efficient breakdown of stained proteins [5,6]. In addition, alkaline proteases are used in numerous other industries (e.g., foods, pharmaceuticals, peptide synthesis industries, etc.) as well as in degradation of proteinaceous waste and extraction of silver from used X-ray film [7–9]. Therefore, there is a pressing need for new sources of alkaline proteases with improved specificity and stability to meet growing industrial demand [10].
The production of alkaline protease by different strains of Pseudomonas aeruginosa is well documented [11]. However, their native protease activity is insufficient to serve industrial purposes [12]. In addition, proteases are prone to inactivation by autolysis due to thermal and operational conditions [13]. Therefore, improvement of protease yielding strains and optimization of cultural parameters are necessary for the enhancement of protease production. Strain improvement by UV exposure is very useful for higher protease yield [14]. Moreover, UV irradiation as a means of random mutagenesis [15–17] and cultural optimization [18–20] for enhancement of protease activity are previously reported. Furthermore, optimization of cultural parameters (e.g., temperature, pH, incubation period, media supplements, etc.) is being used in the industrial process for making enzyme production higher, cost-effective and economically viable [12]. Studies on alkaline protease production by Bacillus spp. [18, 21, 22] and Pseudomonas spp. [23–26] have been reported previously. However, reports on the enhancement of alkaline protease production by P. aeruginosa through UV irradiation and cultural optimization are very few.
The current study was intended to evaluate P. aeruginosa for alkaline protease activity in vitro and generation of potential mutants with hyper-protease activity through UV-induced mutagenesis. In addition, cultural and nutritional optimization of the potential mutant for enhanced protease yield were also studied.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The wild-type P. aeruginosa MZ2F and P. aeruginosa MZ4A strains were obtained from our previous study [27] at the Laboratory of Microbiology, Department of Biotechnology and Genetic Engineering, Islamic University, Bangladesh, in which these bacteria were isolated and identified using the standard protocol. The bacteria were revived from glycerol stock (-80 ºC) by culturing on nutrient agar (NA) plates for 24 h at 37 ºC. The NA plates were maintained at 4 ºC for further investigations.
In vitro determination of protease activity
Qualitative protease activity was determined according to Latorre et al. (2016) with slight modifications [28]. Briefly, selected strains were grown in nutrient broth (NB) at 37 °C for 24 h. To estimate protease activity, 10 μl of each bacterial suspension (0.5 McFarland, 1.5×108 CFU/mL) were placed in the center of the skim milk (SM) agar plate (pH adjusted to 8.0) and incubated for 24 h at 37°C. After incubation, the plates were evaluated by measuring the diameters of the clearance zone and bacterial colony. The relative proteolytic activity (RPA) was determined by using the formula:
Based on RPA value, parent strains were categorized into excellent (RPA > 5.0), good (2.0 ≤ RPA ≥ 5.0), or poor (RPA ≤ 2.0) protease producers.
Quantitative assay of protease activity
The protease activity was estimated by the method described by Beg et al. (2002) with minor modifications [29]. Briefly, following incubation, the bacterial broth was centrifuged at 7000 rpm for 15 min at 4ºC to obtain the cell-free supernatant (CFS). Then 0.5 mL of CFS was added to 1 mL of 1% (w/v) casein solution in the glycine-NaOH buffer of pH 10.0 and incubated for 15 min at 37 ºC. The reaction was discontinued by adding 4 mL of 5% (v/v) trichloroacetic acid (TCA) kept at 25 ºC for 30 minutes. The supernatant was examined by measuring optical density at 660 nm (OD660) using a spectrophotometer (UV mini-1240, Shimadzu, Japan). The liberated tyrosine content of the supernatant was compared with the standard curve of tyrosine. A single unit (U) of protease activity was calculated as the amount of enzyme that released 1 µg tyrosine per min at 37 ºC under assay conditions.
Mutagenesis and selection of potential mutants
The mutagenesis greatly depends on the time and intensity of UV exposure as well as the physiology of bacterial cells [30]. Therefore, wild-type strains were exposed to UV irradiation for different length of period [17]. In brief, bacterial cells were grown in NB medium for 24 h at 37 ºC with shaking. The culture broth was centrifuged at 10,000 rpm for 10 min at 4ºC and precipitated pellets were suspended with 0.9% NaCl. Then, 2 ml of bacterial suspension was placed onto glass Petri dishes (15 cm diameter) and treated with UV radiation (30w, 2537 Å) illuminating at 45º angle at room temperature for different time interval. An untreated plate was considered as a control plate. A portion of treated bacterial strains (100 µL) was spread on SM agar plates and incubated at 37 ºC for 24 h. Colonies developed after incubation were counted and potential mutants were selected based on their protease activity in submerged culture.
Fermentation for protease production
Submerged culture for protease production was performed as described by Singh and Bajaj (2015) [31]. Briefly, wild and mutant strains (0.5 McFarland, 1.5×108 CFU/ml) were cultured in media containing (%, w/v) 1.0 glucose, 0.5 peptone, 0.5 yeast extract, 0.4 K2HPO4, 0.1 Na2HPO4, 0.01 MgCl2, and 0.6 Na2CO3 at pH 9.0. The fermentation was conducted at 37 ºC for 48 h. After incubation, the culture broth was withdrawn and centrifuged, and the supernatant (crude enzyme) was assayed for protease activity as described earlier.
Optimization of physical parameters
The influence of temperature, pH of the medium, and incubation period on alkaline protease production by the selected strain was investigated at varying temperature (viz., 30, 35, 40, 45, and 50 °C), initial pH (viz., 6, 7, 8, 9, 10, and 11), incubation period (viz.,12, 24, 36, 48, 60, and 72 h) while keeping the other parameters constant. The bacterial inoculum (0.5 McFarland, 1.5×108 CFU/ml) was added into the media containing 0.5% yeast extract and 1% glucose as nitrogen and carbon sources, respectively. Finally, the pH of the broth was fixed with 1N NaOH/HCl using a pH meter.
Optimization of medium supplements
Different types of carbon sources (1%, w/v) and nitrogen sources (0.5%, w/v) were added as media supplements to evaluate their effect on protease production. The carbon sources used were sucrose, glucose, lactose, and starch while peptone, beef extract, casein, and yeast extract were used as nitrogen sources. Medium supplemented with 1.5%, 2%, 2.5%, and 3% of NaCl (w/v) was used to evaluate the influence of salt concentration on alkaline protease production. Protease yield was carried out individually after incubation at 37 ºC for 48 h.
Statistical analyses
All experiments were conducted in triplicate and data were presented as means ± standard deviations (mean ± SD). Standard deviations, one way ANOVA, and two-tailed t-tests were calculated with Microsoft Excel 2013. Protease activity was considered significant when p≤0.05, p≤0.01, and p≤0.005. Graphical representations were prepared by Origin Pro v8.0 (Origin Lab Corporation, Massachusetts, USA).
RESULTS
Inherent protease activity of wild-type strains
The RPA has been used to estimate the degree of protease activity. In this study, the P. aeruginosa strain MZ2F (PA-MZ2F) showed an RPA of 2.53±0.04 (significant at P≤0.05) while P. aeruginosa strain MZ4A (PA-MZ4A) provided RPA of 2.12±0.12 (significant at P≤0.01). Despite the considerable differences in their RPA values, both strain PA-MZ2F and PA-MZ4A showed good RPA profiles in vitro as shown in Table 1.
Table 1. Relative proteolytic activity (RPA) of the wild-type bacterial strains.
Selection of UV-induced hyper-proteolytic mutants
UV exposure was applied to both wild-type strains and the survival data obtained after successful UV treatment are presented in Table 2. However, no viable bacterial cells were found after 35 min of UV irradiation. Table 3 shows that most of the mutants from different periods of UV exposure showed no significant difference in protease production than that of the wild strains. Interestingly, UV exposure (2537 Å) for 25 min resulted in a strain mutated from wild strain P. aeruginosa MZ2F (designated as PA-M25) showed higher protease production. In submerged culture, the mutant strain PA-M25 exhibited 612.84±2.50 U/ml of protease as compared to 349.26±2.57 U/ml of the parent strain (Table 3). Thus, mutant PA-M25 provided almost 2-fold more protease yield than the parent strain, hence, considered as the potential hyper-proteolytic mutant.
Table 2. Number of viable colonies after a different length of UV exposure.
Table 3. UV-induced protease activity by mutant and parent strains of P. aeruginosa.
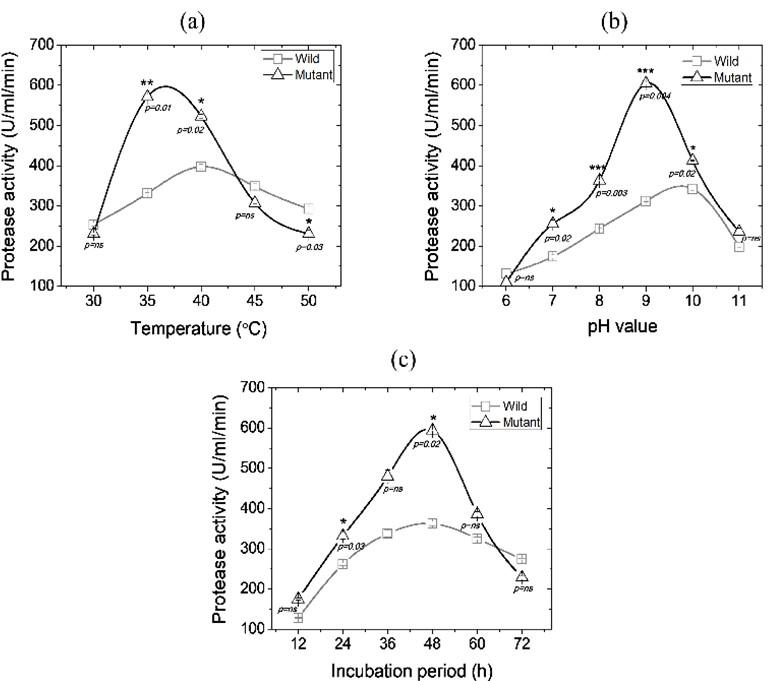
Effect of physical parameters in protease production
The production of microbial enzymes is known to be affected by the physical parameters which directly involved in microbial growth. Figure 1 shows how the temperature, incubation period, and pH influence the protease production by mutant PA-M25. In this study, for instance, mutant PA-M25 showed the highest protease activity at 35 °C which is 571.06±5.63 U/ml while the wild-type strain showed 397.89±5.16 U/ml. The incubation at temperatures other than 35 ºC was found to decrease the protease production (Figure 1a).
In addition to temperature, the pH of the media was found to affect protease activity. The mutant PA-M25 showed maximum protease yield of 605.12±1.98 U/ml at pH 9.0 while wild-type strain exhibited 342.97±0.19 U/ml of protease with pH 10.0 (Figure 1b). We also observed stimulation of enzyme production at alkaline pH followed by a dramatic decrease after pH 9.0. The enzyme production also varies with incubation time [25]. We found an incubation period of 48 h to be the best for protease production for both wild-type strain (362.87±4.09 U/mL) and mutant strain (594.09±5.59 U/ml). However, the production of protease was decreased after 48 h of incubation (Figure 1c).
Effect of media composition on protease production
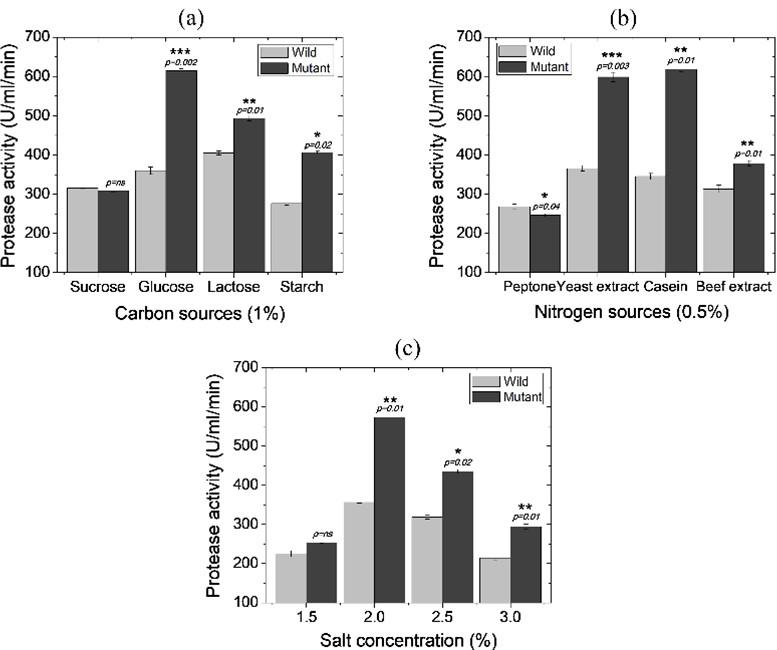
The effects of carbon source, nitrogen source and salinity of the media in protease activity are provided in Figure 2 The highest protease production by wild and mutant strain was achieved with medium containing 1% glucose and 1% lactose, respectively (Figure 2a). With the best carbon supplements, mutant strain PA-M25 gave 615.56±2.79 U/ml of protease production while wild-type strain providing 405.02±1.85 U/ml. Therefore, mutant PA-M25, grown in 1% glucose, provided 1.5-fold more protease production over the wild-type strain. On the other hand, mutant PA-M25 provided the highest protease yield (618.99±1.78 U/ml) with 0.5% casein supplement followed by yeast extract, beef extract, and peptone.
In contrast, maximum protease activity (365.95±3.18 U/ml) by wild-type strain was found with 0.5% yeast extract (Figure 2b). Therefore, casein was evident to be the best nitrogen source for PA-M25 which resulted in 1.7 times more protease activity. Furthermore, 2% NaCl concentration was the best salt concentration for both wild and mutant strains providing 356.37±0.74 U/ml and 573.16±2.83 U/ml of protease production, respectively (Figure 2c). Therefore, mutant PA-M25 showed more than 60% increased protease yield with 2% NaCl than the parent strain.
DISCUSSION
Due to the rapid growth and cultivation with limited resources and ease of genetic manipulation, bacteria-derived alkaline proteases are preferred in industrial applications [32]. In this study, P. aeruginosa strains which showed good RPA profiles were subjected to UV exposure. We found significantly increased protease activity (p≤0.005) in the medium exposed to UV radiation for 25 min by one of the strains. The potential mutant PA-M25 showed 75.47% (~1.75-fold) increased protease production. In a previous study, 10 min of UV radiation generated a mutant from Pseudomonas sp. RAJR 044 which gave 2.5 fold increased protease activity (99.78 U/mL) over the parent strain (40.01 U/mL) when culturing at 35ºC for 48 h [33]. UV-induced enhancement of protease production was also studied for other bacteria as well [17,34]. For instance, Sher et al. (2012) and Wang et al. (2016) found Bacillus subtilis mutants which exhibited 2- and 2.5-fold higher extracellular caseinolytic activity compared to the wild strains, respectively. It is suggested that the degree of enzyme production might increase due to the mutation/damage in the genes reside in bacterial plasmids that might influence the chromosomal genes for alkaline protease production [35].
Bacterial metabolism is affected by the physical parameters of the culture environment. For example, the temperature can influence the secretion of the extracellular enzyme by changing the physical properties of the cell membrane [36]. Therefore, protease production by our mutant PA-M25 is well comparable with several other studies where the bacteria like P. aeruginosa [25,26,37], P. fluorescens [24], B. subtilis [21], Vibrio alginolyticus [38] preferred 35-37 ºC for the highest level of proteolytic activity. In one study, however, B. subtilis showed a high level of protease activity at 45 ºC after 36 h of fermentation with continuous agitation [18]. The pH of the media also known to affect the enzymatic processes and nutrient transportation across the cell membrane [39]. In concurrence to our result, Pseudomonas spp. exhibited the highest protease production at pH 9.0 in earlier studies [24,40]. Similarly, Ash and Ramteke (2018) observed the highest productivity at pH 9.0 using medium supplemented with 1% glucose, 1% yeast extract when cultivated for 36 h at 37 °C. However, pH requirements may vary with bacterial species. For example, maximum protease activity is also found at pH 10.0 by B. subtilis [18]. The hyper-protease activity at alkaline medium provided cue about the alkali nature of the protease. In our study, both mutant PA-M25 and respective parent strain preferred an incubation period of 48 h for maximum yields (P≤0.05). The obtained results coincide with other studies in which protease activity was found maximum after 48 h for P. aeruginosa [25,41]. In another study, P. aeruginosa showed maximum protease activity at pH 9.5, temperature 37 ºC and 48 h of incubation time [42]. Similarly, B. subtilis showed maximum protease yield for an incubation period of 48 h using beef extract [42, 43]. However, protease production from Pseudomonas spp. was found to be maximum at 24 h and decreased afterward unlike the present finding [24,33].
The second most significant parameter regarding protease production is media composition. The mutant PA-M25 provided 1.5 fold more protease production (P≤0.005) over the wild strain with 1% glucose as the carbon source. A similar result was also observed in another study [26]. Additionally, 1% glucose was also suggested as an optimal carbon source for P. aeruginosa in an RSM-based analysis [43]. However, a study demonstrated that both growth and protease production were suppressed when glucose present in the medium [44]. Moreover, increased yields of alkaline proteases were also reported by several studies with other sugar materials including maltose, sucrose, and fructose [25,43,45]. For nitrogen sources, our finding agrees with the findings of other workers as well [46,47]. However, nitrogen sources other than casein have also been reported to give maximum protease yield [24,25,44]. For instance, Patil and Chaudhari (2011) observed higher protease activity (>300.0 U/ml) by P. aeruginosa with beef extract/peptone/yeast extract than with casein (269.3 U/ml) as nitrogen sources when fermentation carried out for 48 h at 40 ºC and pH 9.0 [44]. The osmotic shock (dehydration and/or rehydration) due to NaCl might lead to modifications of the phospholipid structure of the cell membrane that can cause cell death [48]. In our study, 2% of NaCl provided 60% more protease yield than that of wild strain (P≤0.01) which is in line with the findings of other studies [2,38]. Therefore, mutant PA-M25 can be used in industrial applications dealing with high salinity or osmotic pressures. However, we observed a significant reduction (P≤0.05) in protease activity beyond this concentration. This could be due to cell membrane disruption caused by increased salt concentration [2].
CONCLUSIONS
A high protease yielding mutant PA-M25 was generated from wild-type strain P. aeruginosa MZ2F using UV mutagenesis. The mutant PA-M25 exhibited a significant 75.47% (~1.75 fold) more protease yield compared to the parent strain. In addition, no significant damages occurred due to UV irradiation since the morphological features were consistent in both wild and mutant strains. Protease production by PA-M25 reached the maximum level when cultured for 48 h at 35 ºC with medium supplemented with 1% glucose, 0.5% casein and 2% NaCl. Therefore, the mutant PA-M25 is halotolerant and alkaliphilic in nature. These results of the present study demonstrate that strains with higher enzyme activity could be produced with UV mutagenesis. However, further research is needed to characterize the PA-M25 derived alkaline protease and its scale-up fitness before using the mutant in practical applications on a large-scale production.
ACKNOWLEDGEMENTS
This work is financially (partial) supported by Bangladesh University Grant Commission (UGC) research fund fiscal year (2018-19) for experimental expenditure and special thanks to the Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia, Bangladesh for providing laboratory facilities.
CONFLICTS OF INTEREST
Authors declared that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
KR and MMK designed the study; KR, ZN, and BA executed the experimental works; KR and ZN analyzed the data; ZN wrote the manuscript; MMK critically reviewed the manuscript and supervised the whole work. All authors approved the final version of the manuscript.
References
- [1]Bouacem K, Bouanane-Darenfed A, Laribi-Habchi H, Elhoul M Ben, Hmida-Sayari A, Hacene H, et al. Biochemical characterization of a detergent-stable serine alkaline protease from Caldicoprobacter guelmensis. International Journal of Biological Macromolecules. 2015; 81:299–307.
- [2]Ibrahim ASS, Al-Salamah AA, Elbadawi YB, El-Tayeb MA, Ibrahim SSS. Production of extracellular alkaline protease by new halotolerant alkaliphilic Bacillus sp. NPST-AK15 isolated from hyper saline soda lakes. Electronic Journal of Biotechnology. 2015; 18:236–243.
- [3]Raval VH, Pillai S, Rawal CM, Singh SP. Biochemical and structural characterization of a detergent-stable serine alkaline protease from seawater haloalkaliphilic bacteria. Process Biochemistry. 2014; 49(6):955–962.
- [4]Guleria S, Walia A, Chauhan A, Shirkot CK. Purification and characterization of detergent stable alkaline protease from Bacillus amyloliquefaciens SP1 isolated from apple rhizosphere. Journal of Basic Microbiology. 2016; 56(2):138–152.
- [5]Haddar A, Agrebi R, Bougatef A, Hmidet N, Sellami-Kamoun A, Nasri M. Two detergent stable alkaline serine-proteases from Bacillus mojavensis A21: Purification, characterization and potential application as a laundry detergent additive. Bioresource Technology. 2009; 100(3):3366–3373.
- [6]Jain D, Pancha I, Mishra SK, Shrivastav A, Mishra S. Purification and characterization of haloalkaline thermoactive, solvent stable and SDS-induced protease from Bacillus sp.: A potential additive for laundry detergents. Bioresource Technology. 2012; 115:228–236.
- [7]Anwar A, Saleemuddin M. Alkaline proteases: A review. Bioresource Technology. 1998; 64(3):175–183.
- [8]Sathish KR, Ananthan G, Selva PA. Optimization of medium composition for alkaline protease production by Marinobacter sp. GA CAS9 using response surface methodology – A statistical approach. Biocatalysis and Agricultural Biotechnology. 2014; 3(2):191–197.
- [9]Shah K, Mody K, Keshri J, Jha B. Purification and characterization of a solvent, detergent and oxidizing agent tolerant protease from Bacillus cereus isolated from the Gulf of Khambhat. Journal of Molecular Catalysis B: Enzymatic. 2010; 67(1-2):85–91.
- [10]Vijayaraghavan P, Vincent SGP. Cow dung as a novel, inexpensive substrate for the production of a halo-tolerant alkaline protease by Halomonas sp. PV1 for eco-friendly applications. Biochemical Engineering Journal. 2012; 69:57–60.
- [11]Andrejko M, Zdybicka-Barabas A, Janczarek M, Cytryńska M. Three Pseudomonas aeruginosa strains with different protease profiles. Acta Biochimica Polonica. 2013; 60(1):83–90.
- [12]Baweja M, Tiwari R, Singh PK, Nain L, Shukla P. An Alkaline Protease from Bacillus pumilus MP 27: Functional Analysis of Its Binding Model toward Its Applications As Detergent Additive. Frontiers in Microbiology. 2016; 7:1195.
- [13]Anwar A, Saleemuddin M. Alkaline proteases: A review. Bioresource Technology. 1998; 64(3):175–183.
- [14]Beg QK, Saxena R, Gupta R. De-repression and subsequent induction of protease synthesis by Bacillus mojavensis under fed-batch operations. Process Biochemistry. 2002; 37(10):1103–1109.
- [15]Szekeres A, Kredics L, Antal Z, Kevei F, Manczinger L. Isolation and characterization of protease overproducing mutants of Trichoderma harzianum. FEMS Microbiology Letter. 2004; 233(2):215–222.
- [16]Wang HY, Liu DM, Liu Y, Cheng CF, Ma QY, Huang Q, et al. Screening and mutagenesis of a novel Bacillus pumilus strain producing alkaline protease for dehairing. Letters in Applied Microbiology. 2007; 44(1):1–6.
- [17]Sher M, Nadeem M, Syed Q, Irfan M, Baig S. Protease Production from UV Mutated Bacillus subtilis. Bangladesh Journal of Scientific and Industrial Research. 2012; 47(1):69–76.
- [18]Pant G, Prakash A, Pavani JVP, Bera S, Deviram GVNS, Kumar A, et al. Production, optimization and partial purification of protease from Bacillus subtilis. Journal of Taibah University for Science. 2015; 9(1):50–55.
- [19]Singh S, Bajaj BK. Medium Optimization for Enhanced Production of Protease with Industrially Desirable Attributes from Bacillus subtilis K-1. Chemical Engineering Communications. 2015; 202(8):1051–60.
- [20]Sharma A, Gangwar M, Kumar D, Nath G. Phytochemical characterization, antimicrobial activity and reducing potential of seed oil, latex, machine oil and presscake of Jatropha curcas. Avicenna Journal of Phytomedicine. 2016; 6(4):366–375.
- [21]Imtiaz S, Mukhtar H. Production of alkaline protease by Bacillus subtilis using solid state fermentation. African Journal of Microbiology Research. 2013; 7(16):1558–1568.
- [22]Sharma KM, Kumar R, Panwar S, Kumar A. Microbial alkaline proteases: Optimization of production parameters and their properties. Journal of Genetic Engineering and Biotechnology. 2017; 15(1):115–126.
- [23]Izrael-Živković L, Gojgić-Cvijović G, Karadžić I. Isolation and partial characterization of protease from Pseudomonas aeruginosa ATCC 27853. Journal of Serberian Chemical Society. 2010; 75(8): 1041–1052.
- [24]Kalaiarasi K, Sunitha PU. Optimization of alkaline protease production from Pseudomonas fluorescens isolated from meat waste contaminated soil. African Journal of Biotechnology. 2009; 8:7035–7041.
- [25]Raj A, Khess N, Pujari N, Bhattacharya S, Das A, Rajan SS. Enhancement of protease production by Pseudomonas aeruginosa isolated from dairy effluent sludge and determination of its fibrinolytic potential. Asian Pacific Journal of Tropical Biomedicine. 2012; 2(3):S1845–S1851.
- [26]Ash K, Ramteke PW. Optimization of Extracellular Alkaline Protease Production From Pseudomonas aeruginosa Isolated from Soil Samples. International Journal of Agriculture, Environment and Biotechnology. 2018; 11(1):187–194.
- [27]Nain Z, Islam MA, Karim MM. Antibiotic resistance profiling and molecular phylogeny of biofilm forming bacteria from clinical and non-clinical environment in southern part of Bangladesh. International Journal of Enteric Pathogens. 2019; 7(2):37-43.
- [28]Latorre JD, Hernandez-Velasco X, Wolfenden RE, Vicente JL, Wolfenden AD, Menconi A, et al. Evaluation and Selection of Bacillus Species Based on Enzyme Production, Antimicrobial Activity, and Biofilm Synthesis as Direct-Fed Microbial Candidates for Poultry. Frontiers in Veterinary Science. 2016; 3:95.
- [29]Beg QK, Saxena RK, Gupta R. De-repression and subsequent induction of protease synthesis by Bacillus mojavensis under fedbatch operations. Process Biochemistry. 2002; 37:1103-1109.
- [30]Kostyleva EV, Sereda AS, Velikoretskaya IA, Nefedova LI, Sharikov AY, Tsurikova NV, et al. A New Bacillus licheniformis Mutant Strain Producing Serine Protease Efficient for Hvdrolysis of Soy Meal Proteins. Mikrobiologiia. 2016; 85(4):436–445.
- [31]Bajaj BK, Singh NP. Production of xylanase from an alkali tolerant Streptomyces sp. 7b under solid-state fermentation, its purification, and characterization. Applied Biochemistry and Biotechnology. 2010; 162: 1804–1818.
- [32]Najafi MF, Deobagkar D, Deobagkar D. Potential application of protease isolated from Pseudomonas aeruginosa Electronic Journal of Biotechnology. 2005; 8(2).
- [33]Dutta JR, Banerjee R. Isolation and Characterization of a Newly Isolated Pseudomonas Mutant for Protease Production. Brazilian Archives of Biology and Technology. 2006; 49(1):37–47.
- [34]Wang XC, Zhao HY, Liu G, Cheng XJ, Feng H. Improving production of extracellular proteases by random mutagenesis and biochemical characterization of a serine protease in Bacillus subtilis S1-4. Genetics and Molecular Research. 2016; 15(2).
- [35]Solaiman EAM, Wafaa K, Hegazy MEM. Induction of overproducing alkaline protease Bacillus mutants through UV irradiation. Arab Journal of Biotechnology. 2005; 8(1):49–60.
- [36]Abusham RA, Rahman RNZR, Salleh AB, Basri M. Optimization of physical factors affecting the production of thermo-stable organic solvent-tolerant protease from a newly isolated halo tolerant Bacillus subtilis strain Rand. Microbial Cell Factories. 2009; 8:20.
- [37]Zambare V, Nilegaonkar S, Kanekar P. A novel extracellular protease from Pseudomonas aeruginosa MCM B-327: enzyme production and its partial characterization. New Biotechnology. 2011; 28(2):173–181.
- [38]Shanthakumari AR, Nagalakshmi R, Ramesh S. Scaleup and media optimization of protease by Vibrio alginolyticus. Journal of Ecobiotechnology. 2010; 2(2):17–25.
- [39]Moon SH, Parulekar SJ. A parametric study ot protease production in batch and fed-batch cultures of Bacillus firmus. Biotechnology and Bioengineering. 1991; 37(5):467–483.
- [40]Ariole CN, Ilega E. Alkaline protease production by Pseudomonas aeruginosa isolated from the gut of Pila ovata. Journal of Global Science. 2013; 2(5):126-131.
- [41]Vasantha ST, Subramanian AT. Optimization of cultural conditions for the production of an extracellular protease by Pseudomonas International Current Pharmaceutical Journal. 2012; 2(1):1–6.
- [42]Shivakumar S. Co-production of alkaline protease and amylase of Bacillus sp Y in solid state cultivations. Research Journal of Biotechnology. 2012; 7(2):32–37.
- [43]Nejad ZG, Yaghmaei S, Hosseini RH. Production of Extracellular Protease and Determination of Optimal Condition by Bacillus licheniformis. International Journal of Engineering – Transactions B: Applications. 2009; 22(3):221–228.
- [44]Patil U, Chaudhari A. Optimal production of alkaline protease from solvent-tolerant alkalophilic Pseudomonas aeruginosa MTCC 7926. Indian Journal of Biotechnology. 2011; 10:329-339.
- [45]Sevinc N, Demirkan ES. Production of Protease by Bacillus sp. N40 Isolated from Soil and Its Enzymatic Properties. Journal of Biological and Environmental Science. 2011; 5(14):95-103.
- [46]Ahmad V, Kamal A, Ahmad K, Khan MS. Protease characteristics of bacteriocin producing Lysinibacilli, isolated from fruits and vegetable waste. Bioinformation. 2014; 10(1):13–18.
- [47]Sharma AK, Sharma V, Saxena J, Yadav B, Alam A, Prakash A. Optimization of protease production from bacteria isolated from soil. Applied Research Journal. 2015; 1(7):388–394.
- [48]Sochocka M, Boratyński J. Osmoregulation – an important parameter of bacterial growth. Postepy Higieny I Medycyny Doswiadczalnej. 2011; 65:714–724.